Method for detecting copy number variation of tumor single-cell genomes
A single-cell and genome technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as allele loss, ratio deviation, and inability to eliminate deviations, achieving good results, improving accuracy, and stability Effect of copy number numerical regression
Active Publication Date: 2019-07-16
PEKING UNIV
View PDF1 Cites 7 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Regardless of the single-cell amplification technology, there will be a certain Allele DropOut (ADO) ratio, and the deviation of the allele amplification ratio and the regional amplification preference greatly limit the resolution of single-cell genome copy number analysis. and accuracy
[0005] Existing methods for analyzing genome-wide copy number changes in bulk cells are not well suited for single-cell studies
However, there are certain problems with single-cell genome copy number change analysis methods.
For example: the method of using dynamic interval window plus GC content correction cannot eliminate some specific biases introduced by different single-cell amplification technology methods; and the single-cell genome copy number analysis method of hidden Markov model often appears overfitting ( overfitting), or the case where the degree of fitting is not high
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
[0033] The following is a more detailed implementation description of the present invention. The parameters and specific implementation details are used to explain the feasibility and implementation effects of the present invention, and do not constitute a limitation to the present invention.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
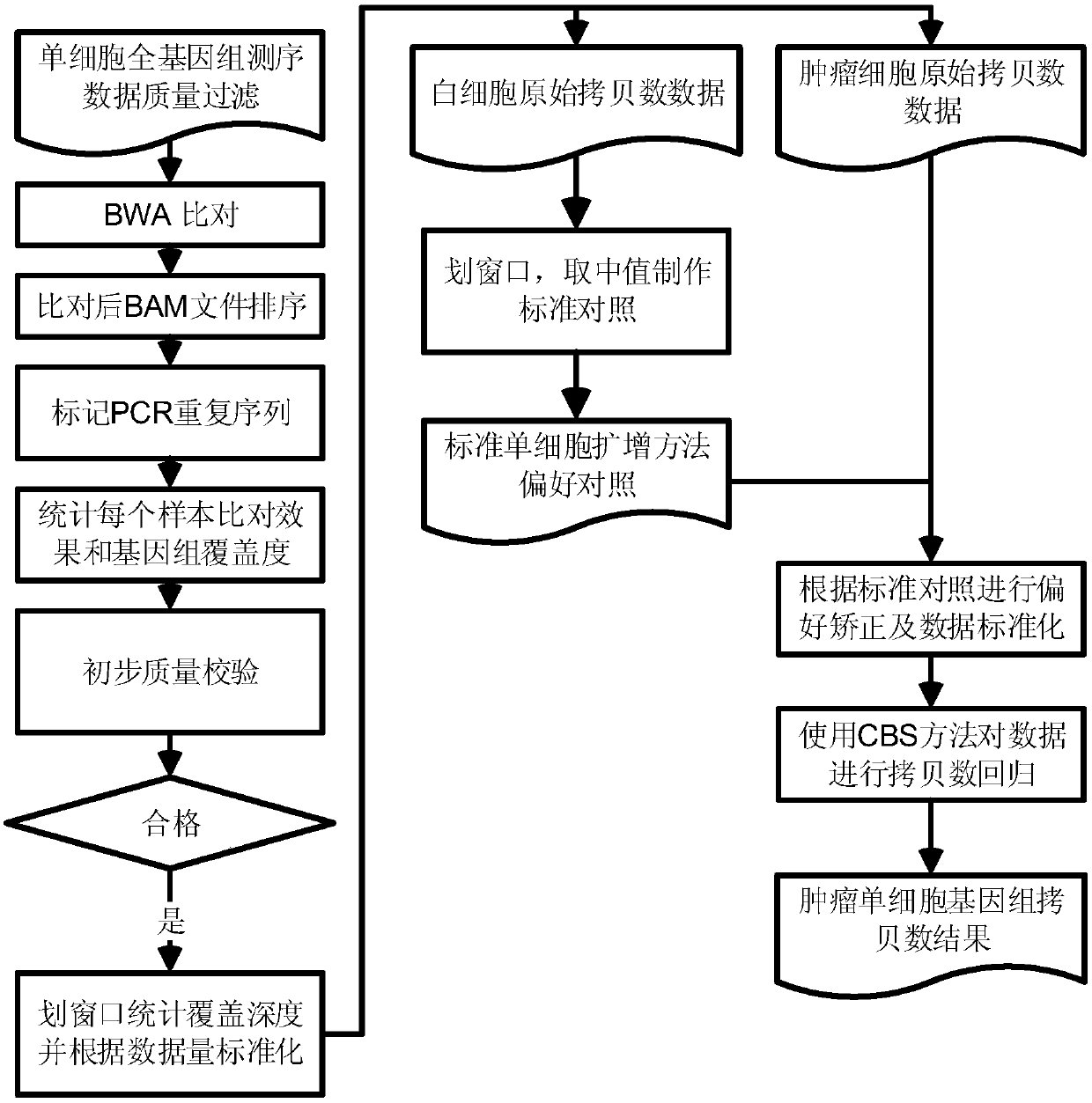
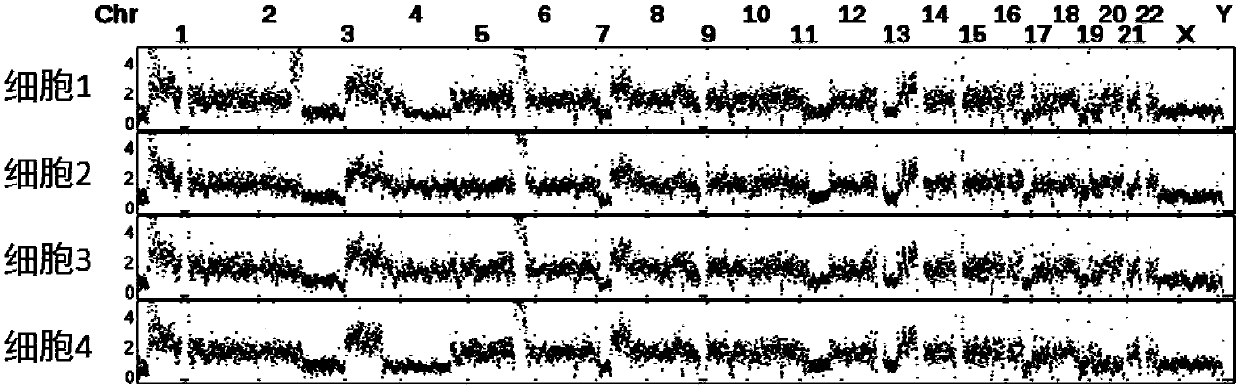
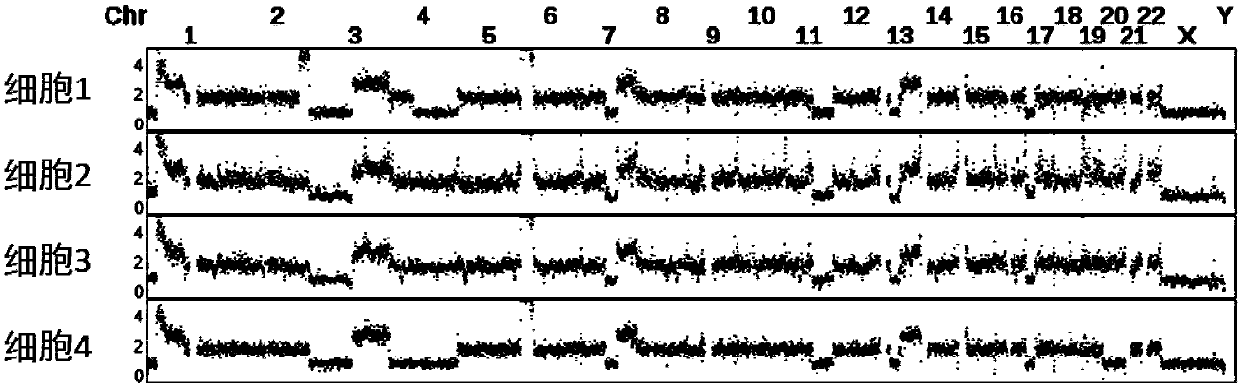
The invention discloses a method for detecting copy number variation of tumor single-cell genomes. The method comprises the steps of separately performing genome sequencing on tumor single cells and aplurality of normal tissue single cells, performing comparing with a reference genome, and dividing window and standard window data; and using a normal single-cell comparison as a standard for removing systematic deviation to obtain genome copy number in all tumor single-cell regions. The method disclosed by the invention eliminates some specific system deviation introduced by different single-cell amplifying techniques, so that the accuracy of copy number variation detection of the tumor single-cell genomes can be improved, and the method has important significance in detecting driving incidents relevant with copy number variation and reading tumor genome evolution information.
Description
Technical field [0001] The invention relates to the fields of single-cell genome sequencing, tumor genome analysis and bioinformatics, in particular to a method for detecting the copy number status of tumor single-cell genome based on second-generation sequencing technology. Background technique [0002] Genome sequencing technology has been widely used in basic research in life sciences and some corresponding translational science applications. At present, the second-generation short-sequence sequencing technology based on solexa sequencing technology has demonstrated its wide range of application and huge application prospects. Based on different purposes, we can directly sequence DNA to assemble new species or detect changes in the genome of species with existing reference sequences, such as single nucleotide polymorphism sites (SNPs, singlenucleotide polymorphism), short sequence insertions or deletions (INDEL, insertion and deletion), genome structural variation (SV, struct...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/6869
CPCC12Q1/6869G16B20/00G16B30/00
Inventor 苏哲倪晓晖高妍白凡
Owner PEKING UNIV
Who we serve
- R&D Engineer
- R&D Manager
- IP Professional
Why Patsnap Eureka
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com