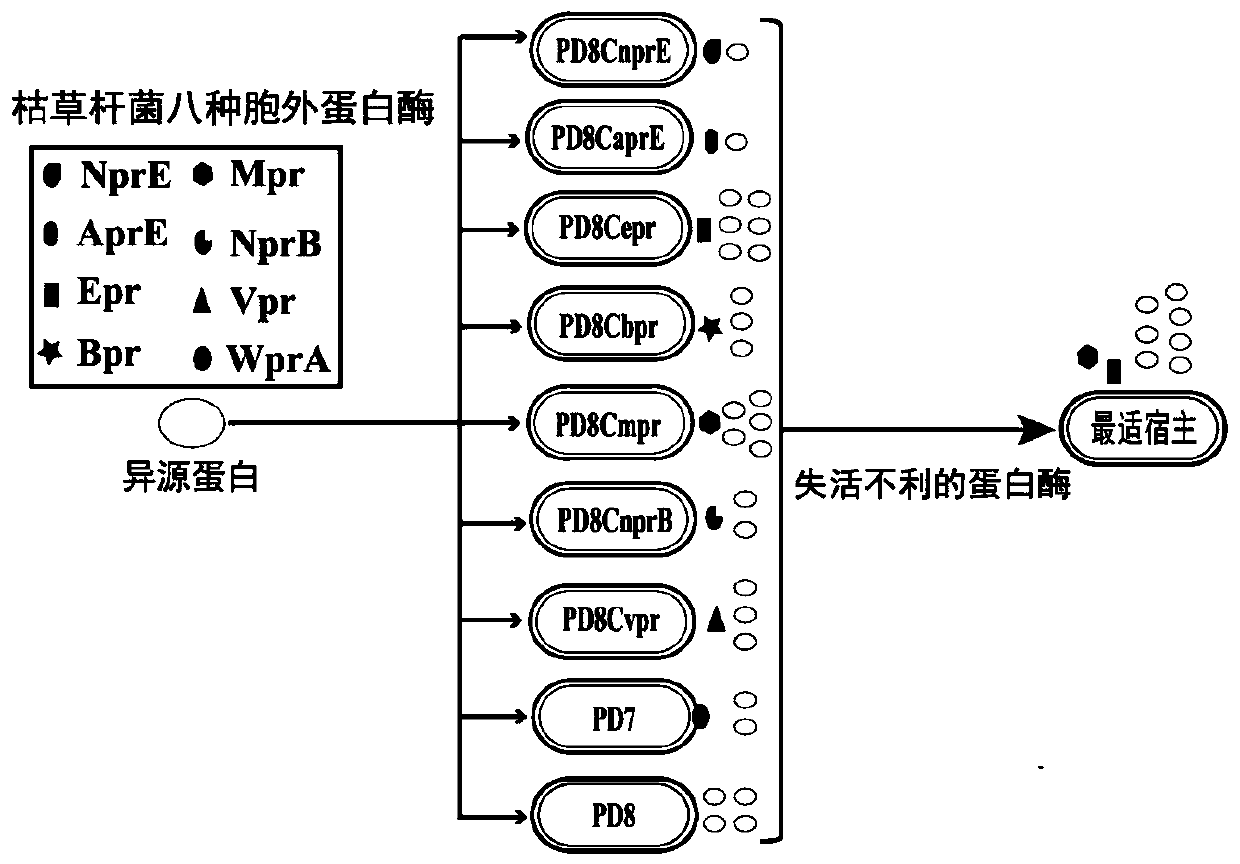
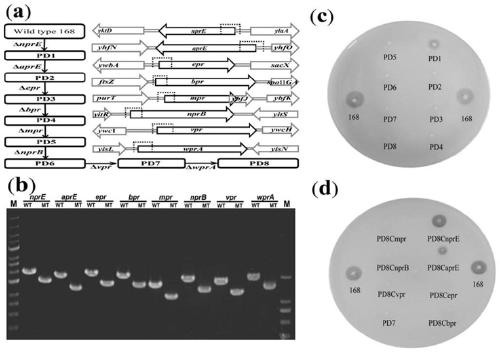
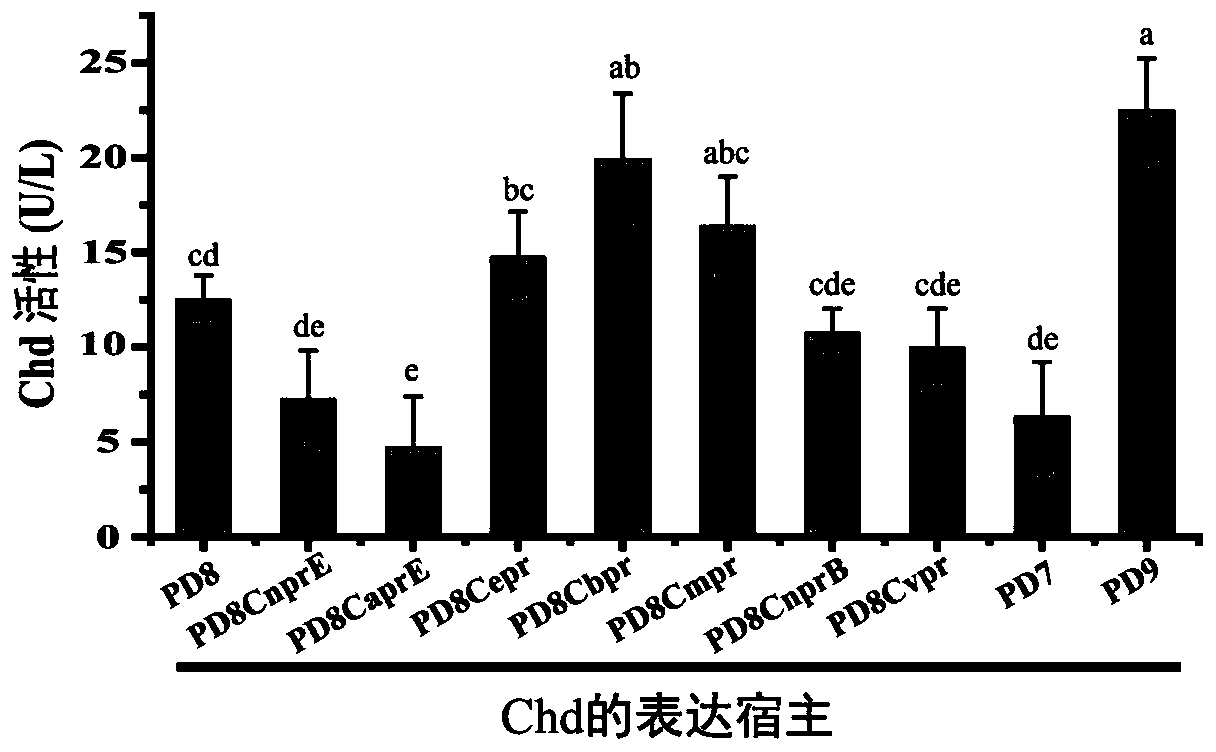
Method for cooperatively establishing optimal bacillus subtilis protease deletion expression host for target protein
A technology of Bacillus subtilis and Bacillus subtilis, applied in the field of biological high, can solve the problems of inability to meet individual needs and poor applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 9
[0018] Example 1 Construction of nine strains of subtilisin deletion mutants
[0019] Such as figure 2 As shown in a and 2b, using Bacillus subtilis 168 as the starting strain, use the pheS* reverse selection marker (ApplMicrobiol Biotechnol, 2017, doi:10.1007 / s00253-016-7906-9) to delete nprE, aprE, epr, bpr, mpr in sequence , nprB, vpr and wprA eight extracellular protease genes to construct Bacillus subtilis mutant PD8. Taking the deletion of nprE as an example, using the total DNA of Bacillus subtilis 168 as a template, using the primer pair nprE-LF-F / nprE-LF-R (SEQ ID NO.1 / SEQ ID NO.2), nprE-RF-F / nprE- RF-R (SEQ ID NO.3 / SEQ ID NO.4) and nprE-DR-F / nprE-DR-R (SEQ ID NO.5 / SEQ ID NO.6) respectively amplify the upstream homology arm (upstream homologous arm, UHA), direct repeat sequence (direct repeat sequence, DR) and downstream homologous arm (downstream homologous arm, DHA), while using the primer pair nprE-PC-F / nprE-PC-R to plasmid pTPC (Appl Microbiol Biotechnol, 2017...
Embodiment 2
[0021] Example 2 Determining beneficial and harmful extracellular proteases to the expression of chlorothalonil hydrolysis dechlorinase (Chd)
[0022] The chlorothalonil hydrolysis dechlorinase gene chd (NCBI sequence number: GQ292539) was connected to the vector pP43NMK (ApplEnviron Microbiol, 2005, doi: 10.1128 / aem.71.7.4101-4103, NCBI sequence number DQ264732) to obtain the expression vector pP43Chd. The specific construction process is as follows: using the total DNA of the bacterial strain Pseudomonas sp.CTN-3 (JBacteriol, 2010, doi: 10.1128 / JB.01547-09) as a template, use the primer pair chd-F / chd-R (SEQ IDNO.9 / SEQ ID NO.10) to amplify chd; the amplified product was ligated with pP43NMK digested by HindⅢ and PstI in vitro by homologous recombination, and the ligated product was transformed into Escherichia coli DH5α competent cells. Transformants were screened on LB plates and verified by sequencing to obtain pP43Chd. Transform pP43Chd into the above-mentioned nine mut...
Embodiment 3
[0023] Example 3 Determining beneficial and harmful extracellular proteases for recombinant expression of amylase AmyM
[0024] The amylase gene amyM (NCBI sequence number KM114206) was connected to the vector pP43NMK to obtain the expression vector pP43AmyM. The specific construction process is as follows: using the total DNA of the strain Corallococcus sp.EGB (Appl EnvironMicrobiol, 2018, doi: 10.1128 / AEM.00152-18) as a template, using the primer pair amyM-F / amyM-R (SEQID NO.11 / SEQ ID NO.12) amplified amyM, ligated the amplified product with pP43NMK digested by HindⅢ and PstI in vitro homologous recombination, transformed the ligated product into E. Transformants were screened on plates and verified by sequencing. Transform pP43AmyM into the above-mentioned nine mutant strains of Bacillus subtilis (PD8, PD7, PD8CnprE, PD8CaprE, PD8Cepr, PD8Cbpr, PD8Cmpr, PD8CnprB and PD8Cvpr), and ferment and culture the obtained nine expressing strains respectively. The medium used is Supe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com