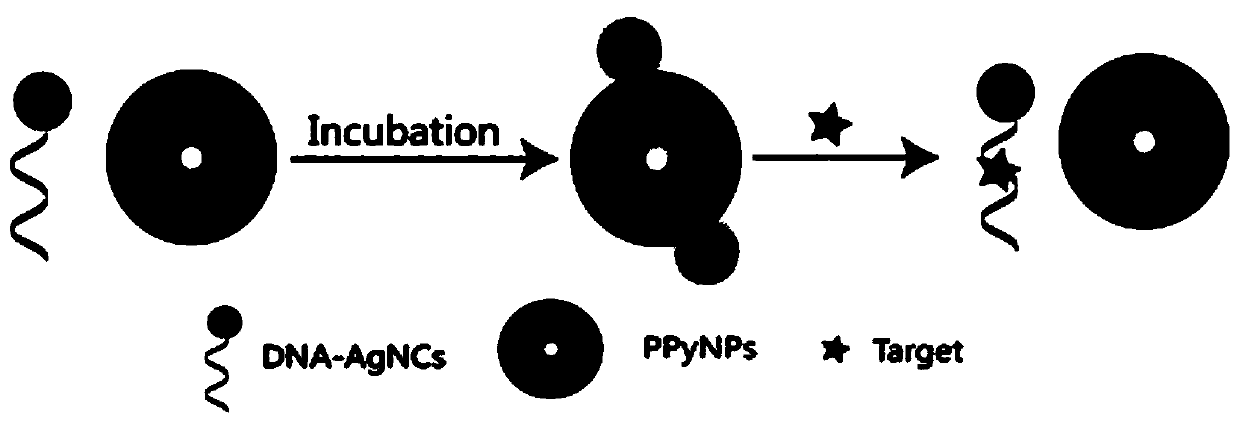
Method for detecting multiple targets based on nucleic acid aptamer fluorescence sensor
A fluorescent sensor and nucleic acid aptamer technology, applied in the field of analysis and detection, can solve the problems of low sensitivity, time-consuming detection, cumbersome steps, etc., and achieve the effect of low cost, strong versatility, and simple detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] A method for detecting target molecules such as interferon-γ, adenosine, and thrombin, comprising the following steps:
[0042] 1) Synthesis of DNA-AgNCs: Dissolve 1OD AgP1 template in 975 μL buffer 2, heat to 95°C, maintain for 10 min, then quickly transfer to an ice-water bath, rapidly cool down, then add 12 μL, 10 mM fresh silver nitrate solution, vortex After swirling and mixing, react at 4°C for 30 minutes in the dark, then quickly add 12 μL of 10 mM freshly prepared sodium borohydride solution, shake well and mix for 1 min, then react overnight at 4°C in the dark; dissolve 1OD of AgP2 template in 975 μL of buffer 2 , heated to 95°C, maintained for 10min, then quickly transferred to an ice-water bath, cooled rapidly, then added 16μL, 10mM freshly prepared silver nitrate solution, vortexed and mixed, and reacted at 4°C for 30min in the dark, then, quickly added 8μL, 10mM freshly prepared silver nitrate solution Sodium solution, shake and mix well for 1 min, then rea...
Embodiment 2
[0055] Selective experiment of detection method of the present invention
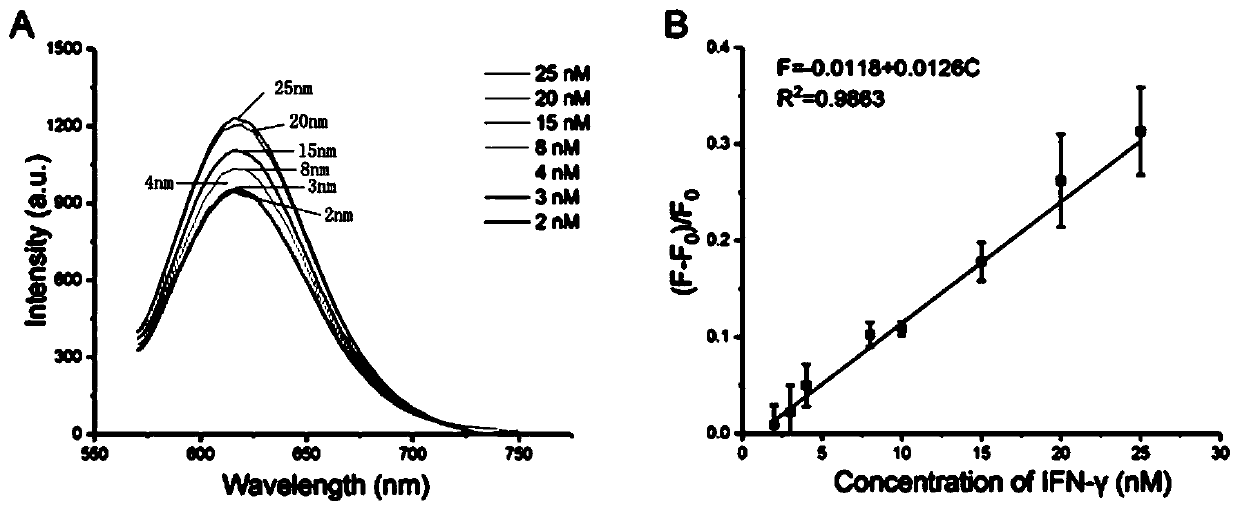
[0056] Repeat steps 1)-step 3) in Example 1, and replace the interferon-γ in step 3) with interfering substances such as bovine serum albumin, lysozyme, albumin, dopamine, L-cysteine, With other conditions unchanged, the fluorescence is detected, and the selectivity result diagram of the method for detecting the target interferon-γ is obtained, as shown in Figure 4 As shown, it can be seen from the figure that the method of the present invention has good selectivity to the target interferon-γ.
Embodiment 3
[0058] The recovery rate experiment of detection method of the present invention
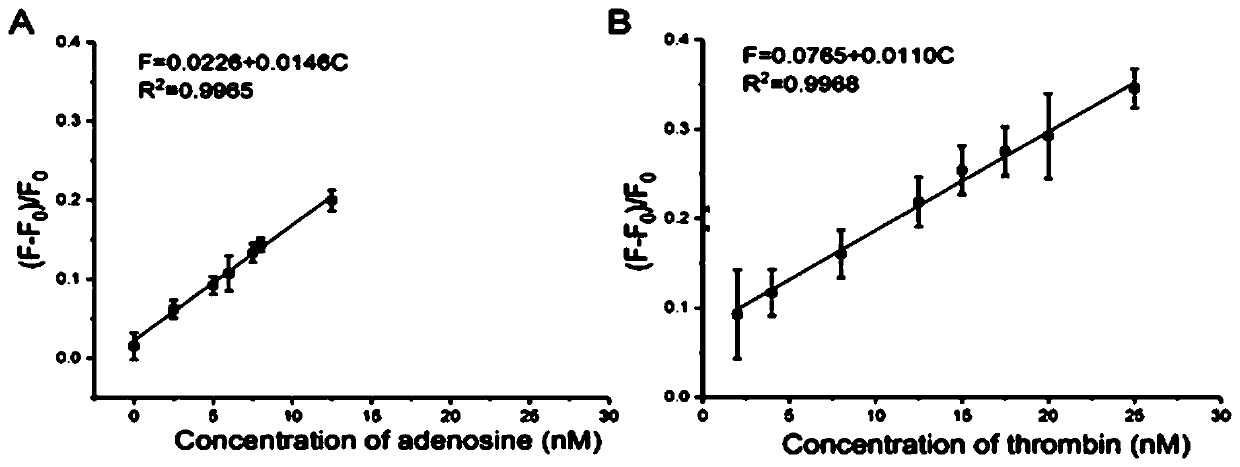
[0059] Repeat step 1)-step 4) of 1) in the embodiment, in step 4), carry out adding sample detection respectively, add the adenosine of 7.8, 9.8, 11.8nM respectively, add the thrombin of 8, 10, 12nM or add 4.5, 6, 7.5nM interferon-γ was detected, and the recovery rate of this method in the actual sample detection was obtained. The results are shown in Table 2.
[0060] Table 2 Using this method to detect the recovery rate of adenosine, thrombin, and interferon-γ in serum samples
[0061]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com