FISH probe set for detecting NTRK fusion and application thereof
A probe group and probe technology, applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of continuous activation of kinase region, abnormal expression of TRK, low expression level, etc. , improve the quality of life, the effect of high probe specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
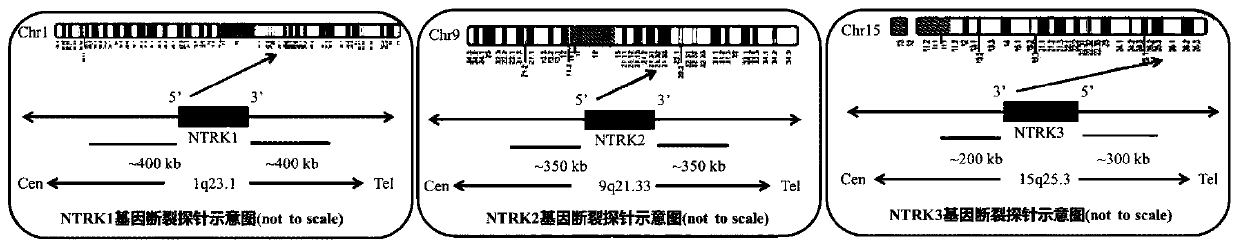
[0042] Such as figure 1 As shown, the NTRK1 5'-end probe set (NTRK1 gene centromere side) was based on two BAC clones (RP11-1054F3 and RP11-66D17) as templates, and the NTRK1 3'-end probe set (NTRK1 gene telomeric side ) with 2 BAC clones (RP11-1038N13 and RP11-110J1) as templates; NTRK2 5' end probe (NTRK2 gene centromere side) with 2 BAC clones (RP11-324L15 and CTD-2509D1) as templates, The NTRK2 3' end probe (NTRK2 gene telomere side) uses 2 BAC clones (RP11-152D13 and CTD-2339E11) as templates; NTRK3 5' end probe (NTRK3 gene telomere side) uses 2 BAC clones (RP11-110O23 and RP11-97O12) were used as templates, and the NTRK3 3' end probe (centromere side of NTRK3 gene) used two BAC clones (RP11-113C11 and CTD-2526N1) as templates. The 5′-end probes of the three genes are all labeled with FITC (the first fluorescent reporter group) as green signals, and the 3’-end probes are all labeled with Tetramethyl-rhodamine (the second fluorescent reporter group) as red signals. The ab...
Embodiment 2
[0062] The steps of using a UV spectrophotometer to measure the labeling efficiency and concentration of the probes in the above-mentioned probe group are as follows:
[0063] (1) Turn on the UV spectrophotometer (NanoDrop), select the full-wavelength mode, and select the corresponding wavelength. After the blank calibration, measure the A2 of the 5′-end probe group respectively. 60nm 、A Dye (FITC:A 494nm ) and A of the 3′ end probe set 260nm 、A Dye (Tetramethyl-rhodamine: A 560nm ).
[0064] (2) Since the fluorescent dye also has a certain absorbance value at 260nm, in order to make the absorbance value of the probe at 260nm more accurate, use the following formula to correct A 260 Value: A Base =A 260 -(A Dye ×CF 260 ), CF 260 Refers to the correction factor, which is a constant, and each fluorescent dye has a certain CF 260 Values, such as the CF of Tetramethyl-rhodamine 260 The value is 0.06 and FITC is 0.32. (3) Calculate the probe labeling efficiency and con...
Embodiment 3
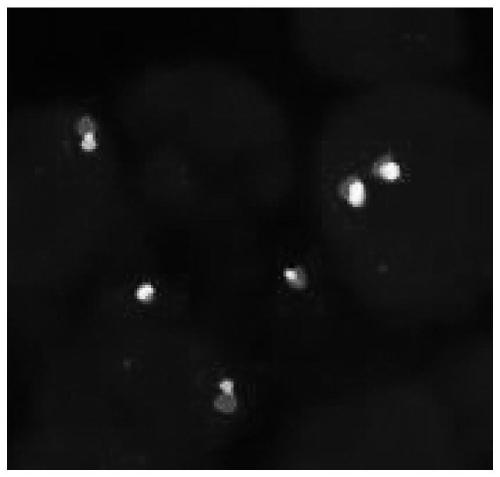
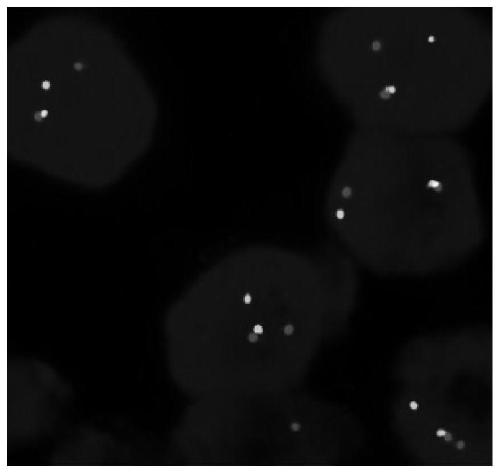
[0069] Using the NTRK1 gene break probe system and the NTRK3 gene break probe system prepared in Example 1 to detect 50 cases of papillary thyroid cancer samples provided by the hospital, the NTRK3 gene break probe system to detect 10 cases of salivary gland mammary secretory carcinoma samples, NTRK2 The gene breakage probe system was used to detect 20 samples of pontine glioma, and the NTRK probe of Empire Genomics was used to detect these samples at the same time. The detection steps are as follows:
[0070] (1) Sample dewaxing and rehydration: immerse tissue section samples in 3 tanks of xylene for 10 minutes each time for dewaxing, and then soak in 100%, 100%, 100%, 85% and 70% ethanol for 3 minutes, Finally, immerse in deionized water for 3 min.
[0071] (2) Sample pretreatment and digestion: Boil the tissue section sample in 100°C water for 20 minutes, take out the sample and digest it with pepsin for 10 minutes, transfer it to 2×SSC solution for 3 minutes, take out the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com