Molecular detection method for rapid quantification of diatom cell density
A molecular detection and diatom technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of relying on professional technicians and high-precision instruments, lack of specific quantitative detection methods, and heavy analysis tasks. , to achieve the effect of improving efficiency, simple and convenient operation method, accurate and objective data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Primer design and specificity verification:
[0028] A pair of specific primers rbc280-F / rbc280-R was designed for diatom rbcL gene:
[0029] rbc280-F: 5'-ACGTTTAGAAGATATGCGTATTC-3'
[0030] rbc280-R: 5'-GGTTAATACCTTCCATACAG-3'.
[0031] The primers rbc280-F / rbc280-R were used to verify the existing algae strains, and the results are shown in Table 1.
[0032] Table 1 Verification results of some algae strains
[0033]
[0034]
[0035] Note: "+" means positive detection, "-" means negative and not detected.
[0036] Table 1 uses specific primers to detect the existing algae strains cultivated in the laboratory, including 7 species of cyanobacteria, 14 species of green algae and 9 species of diatoms. From the test results, the detection rate of diatoms in this research method is 100%. , but not detected for cyanobacteria and green algae, showing the specificity of this primer.
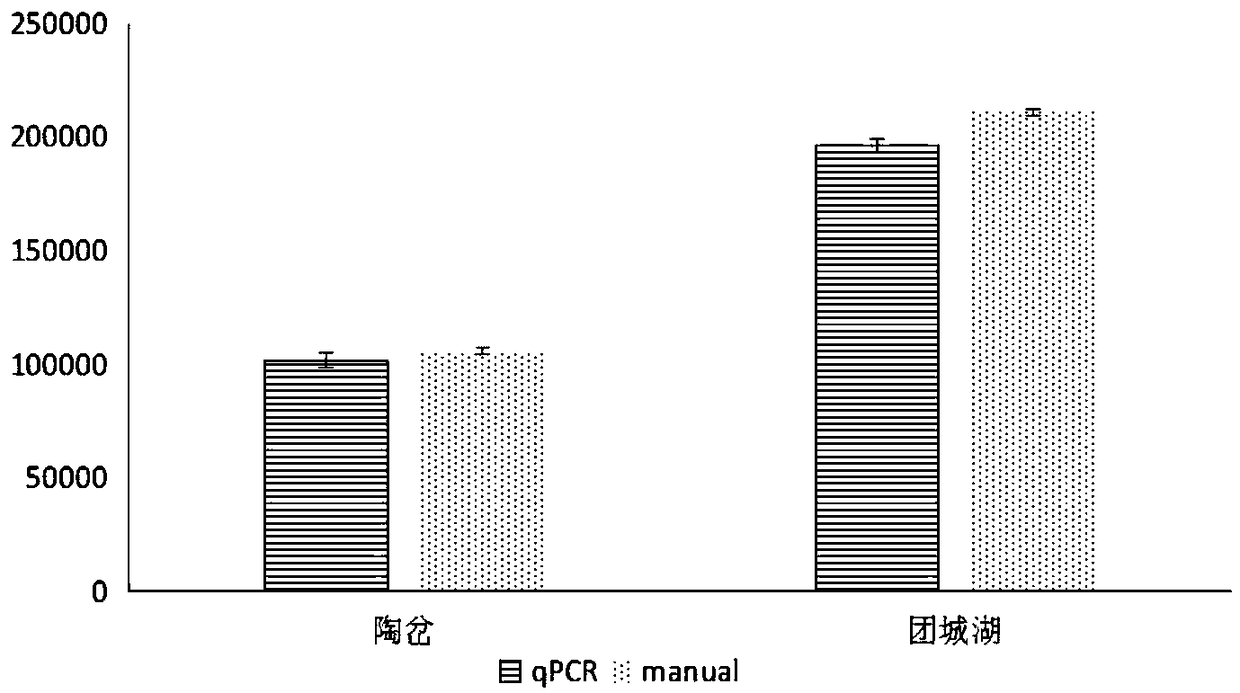
[0037] figure 1 The method is used to detect field samples, and the detection ra...
Embodiment 2
[0039] A molecular detection method for rapidly quantifying diatom cell density, the steps of which are:
[0040] 1. Sample collection and DNA extraction:
[0041] In April 2018, water samples from the main channel of the South-to-North Water Transfer Project were collected for the quantification of diatoms, and there were 2 sampling points. In this example, 1 L of the water sample to be tested was collected according to the specific conditions of the algae biomass in the water body, and the water sample was filtered through a 0.22 μm cellulose acetate membrane, and the filtered sample was subjected to extraction and purification of total genomic DNA. The extracted DNA was dissolved in sterile double distilled water or deionized water, and stored at -20°C for later use.
[0042] 2. Fluorescence quantitative PCR
[0043] Specific diatom rbcL gene primers were used to amplify and quantify by fluorescent quantitative PCR. The target product size was 280 bp. After the reaction w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com