Nucleic acid purification method for DNA methylation analysis of human-derived stool
A technology for methylation and nucleic acid samples, applied in the field of obtaining nucleic acid samples for DNA methylation analysis, to achieve the effect of quick and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
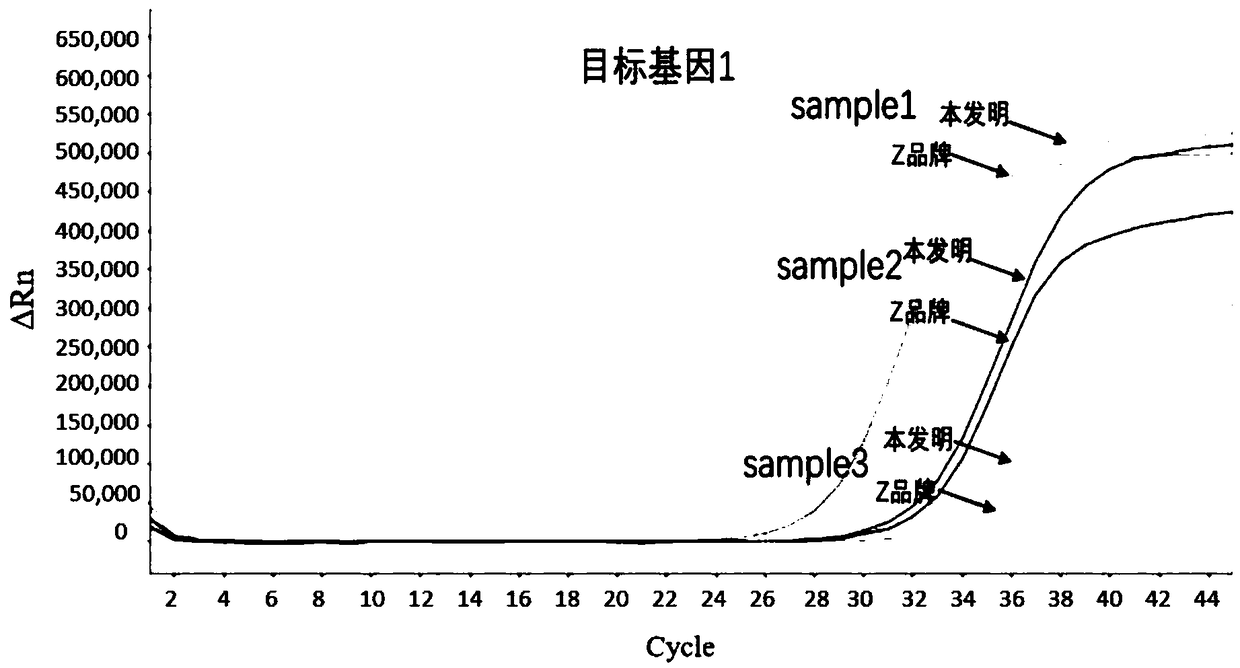
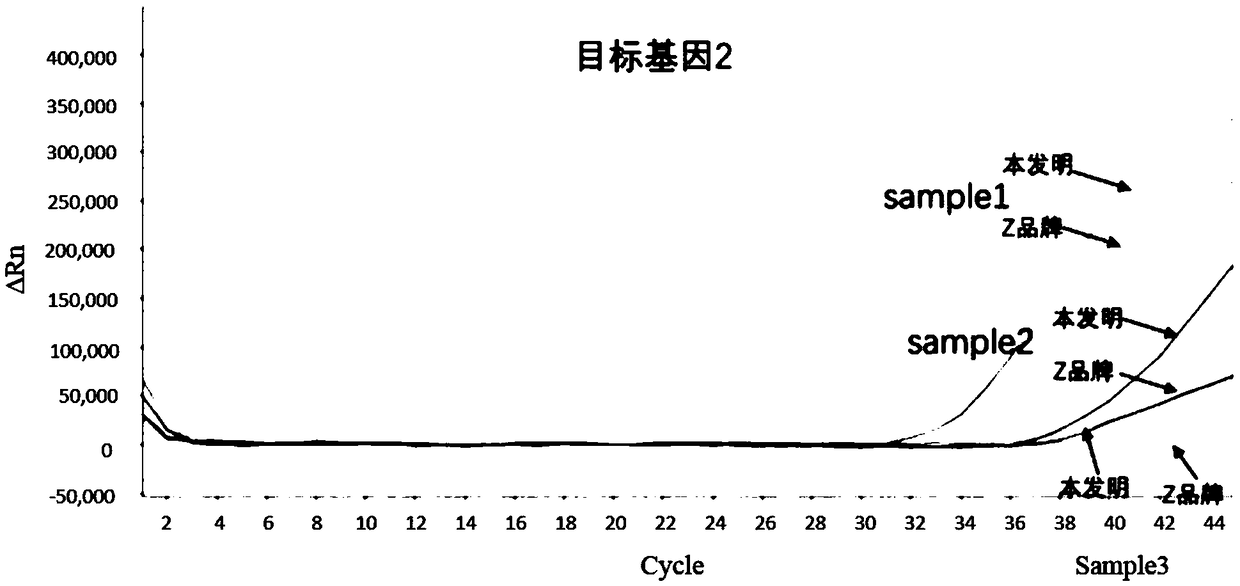
[0067] Example 1 Comparison of the method of the present invention for total human fecal DNA and the method of Z brand commercial sulfite conversion kit
[0068] Obtaining the total DNA of human feces: select several stool samples from patients with colorectal cancer, and obtain the highly methylated target gene through the nucleic acid extraction kit (produced by Shanghai Ruiyi Biotechnology Co., Ltd., Huminxie No. 20180535) Total DNA of human feces.
[0069] 1. The specific implementation steps of the nucleic acid purification method for DNA methylation analysis of the present invention:
[0070] (1) Add 2μg of DNA to be processed (sample1, 2, 3) into a 2.0mL centrifuge tube, and control the input volume range to 20-100μL;
[0071] (2) After adding 190μL of sulfite conversion solution and 30μL of protection solution to the centrifuge tube, turn it upside down to mix, and centrifuge immediately;
[0072] (3) Seal the centrifuge tube in a constant temperature shaking incubator and incu...
Embodiment 2
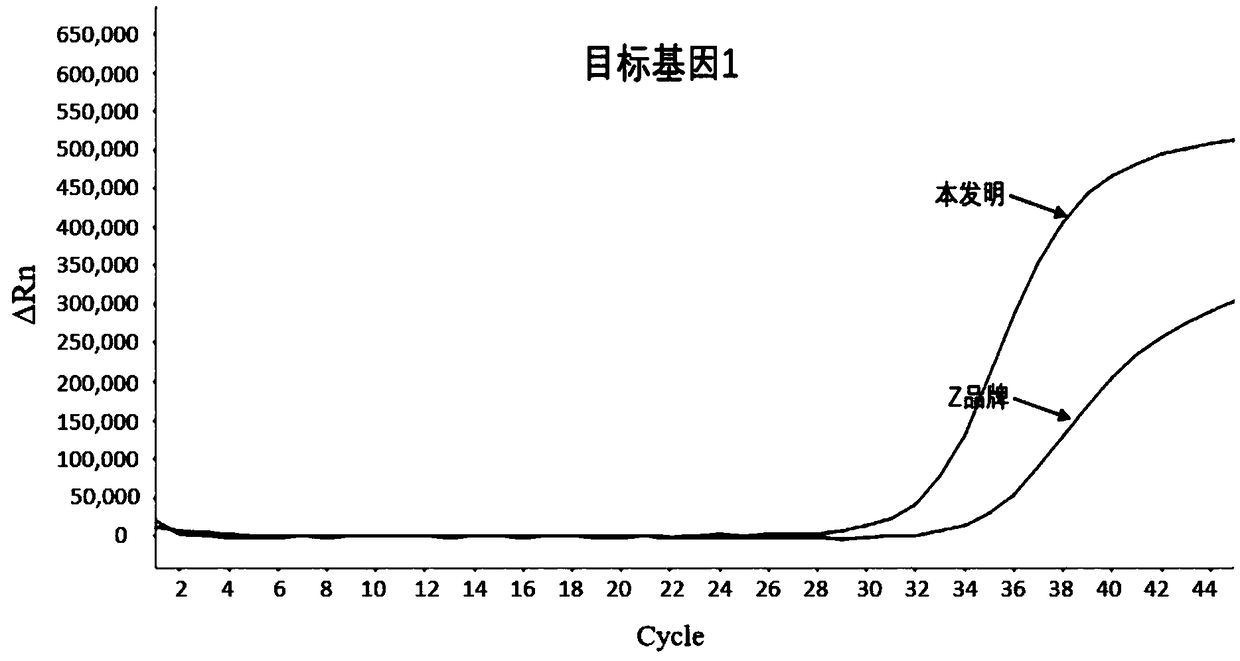
[0109] Example 2 Comparison of the method of the present invention of paraffin DNA and the method of Z brand commercial sulfite conversion kit
[0110] Paraffin-embedded tissue DNA extraction kit ( DNA FFPE Tissue, 56404) to obtain FFPE-DNA.
[0111] 1. The specific implementation steps of the nucleic acid purification method for DNA methylation analysis of the present invention:
[0112] (1) Add 500ng of FFPE-DNA to be processed into a 2.0mL centrifuge tube, the input volume is about 50μL;
[0113] (2) After adding 200μL of sulfite conversion solution and 50μL of protection solution to the centrifuge tube, turn it upside down to mix, and centrifuge immediately;
[0114] (3) Seal the centrifuge tube and place it in a constant temperature shaking incubator, and incubate at 95°C for 5 minutes; to prevent DNA renaturation, immediately remove the centrifuge tube and place it on ice for 5 minutes;
[0115] (4) Take out the centrifuge tube, mix it upside down, centrifuge immediately, place th...
Embodiment 3
[0152] Example 3 Comparison of the method of the present invention for plasma free DNA and the method of Z brand commercial sulfite conversion kit
[0153] Plasma DNA was extracted using peripheral blood free DNA extraction kit (VAHTS Serum / Plasma CircμLating DNA Kit, N902-01) to obtain cfDNA.
[0154] 1. The specific implementation steps of the nucleic acid purification method for DNA methylation analysis of the present invention:
[0155] (1) Add 20ng of DNA to be processed into a 2.0mL centrifuge tube, the volume is 50μL;
[0156] (2) After adding 280 μL of sulfite conversion solution and 50 μL of protection solution to the centrifuge tube, turn it upside down to mix, and centrifuge immediately;
[0157] (3) Seal the centrifuge tube and place it in a constant temperature shaking incubator, and incubate at 95°C for 5 minutes; to prevent DNA renaturation, immediately remove the centrifuge tube and place it on ice for 5 minutes;
[0158] (4) Take out the centrifuge tube, mix it upside do...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com