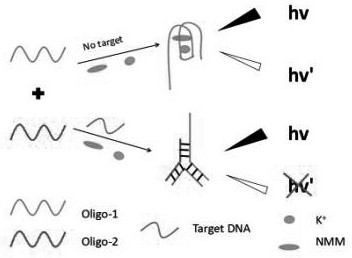
A closed dna fluorescent biosensor and its application in detecting influenza A h1n1 virus
A biosensor and influenza virus technology, applied in the field of biochemical analysis, can solve the problems of false negatives, unsuitable for large-scale screening and detection at the grassroots level, and achieve the effects of easy portability, convenient standardized operation, and low process cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1 Detection of H1N1
[0068] (1) Add 50 μL Oligo-1 (12 μM), 50 μL Oligo-2 (10 μM), 50 μL KCl (500 mM) and 50 μL of target single-stranded DNA (Oligo-3) or samples to be tested in different series of concentrations, and a blank control group was set at the same time (the buffer solution without sample to be tested, only signal probe, auxiliary probe and potassium ion solution was used as blank control group), heated to 88°C for 10 minutes after mixing;
[0069] (2) Gradually cool down to room temperature to form closed double-stranded DNA; then add 50 μL of NMM (48 μM) to the above solution, and incubate at room temperature (20-28°C) for 15 minutes;
[0070] (3) Transfer the above solution to a fluorescence detector, and measure the difference in fluorescence intensity between the experimental group and the blank control group at room temperature; calculate the regression equation according to the standard curve of target single-stranded DNA concentration-fluore...
Embodiment 2
[0074] Embodiment 2 Fluorescence intensity varies with the ratio of Oligo-1 and Oligo-2
[0075] 1. Method
[0076] On the basis of Example 1, the mol ratio of Oligo-1 and Oligo-2 is used as a single factor variable, and ΔI is used as an index (ΔI is defined as the difference of fluorescence intensity, and the definition formula of ΔI is ΔI=I blank -I target , I target A fluorescent signal indicating the presence of the target strand, I blank Indicates that no target strand is added to the fluorescent signal), and the influence of different ratios of Oligo-1 and Oligo-2 on the detection effect of H1N1 virus-specific single-stranded DNA sequences was investigated.
[0077] 2. Results
[0078] Experimental results such as figure 2 As shown, the auxiliary strand (Oligo-2) can hybridize with Oligo-1 and the target strand (Target DNA), and inhibit the formation of G-quadruplexes. The change of the ratio of Oligo-1 and Oligo-2 significantly affects the intensity of the detecte...
Embodiment 3
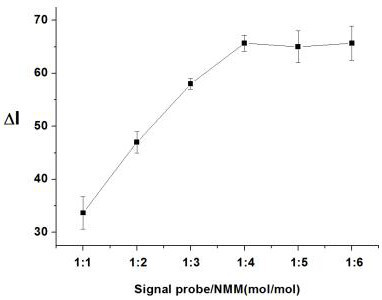
[0079] Embodiment 3 Fluorescence intensity changes with the ratio of Oligo-1 and NMM
[0080] 1. Method
[0081] On the basis of Example 1, the ratio of Oligo-1 and NMM was used as a single factor variable, and ΔI was used as an index to investigate the influence of different ratios of Oligo-1 and NMM on the detection effect of H1N1 virus-specific single-stranded DNA sequences.
[0082] 2. Results
[0083] Experimental results such as image 3 As shown, the fluorescent dye NMM must be combined with the G-quadruplex in a certain ratio to achieve the maximum fluorescent signal value. Depend on image 3It can be seen that when the molar ratio of the signal probe to the fluorescent dye is 1:1-6, the detection effect is better; the fluorescence intensity of the detection system increases gradually with the ratio of Oligo-1 to NMM, and when the G-quadruplex When the molar ratio of probe to NMM is 1:4, the net increase ΔI of the fluorescence signal value of the detection system r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com