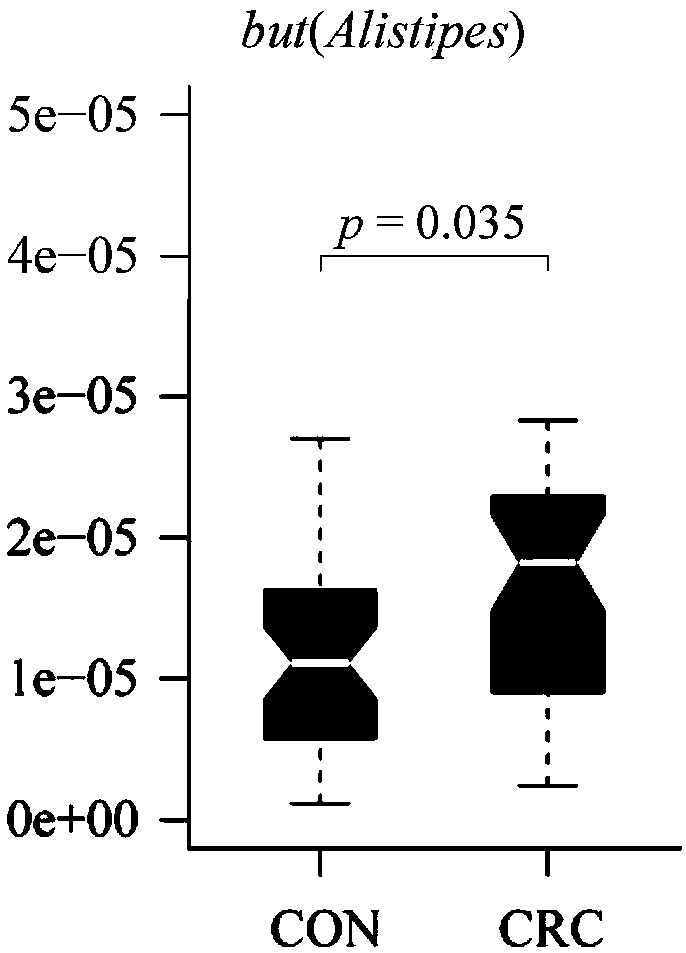Primer pair and kit for detecting butyric acid synthetic gene
A technique for synthesizing genes and primer pairs, which is applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc. It can solve problems such as limited sensitivity and specificity, high incidence of CRC, and hidden early symptoms , to achieve the effect of improving prediction rate, high sensitivity and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Real-time fluorescence quantitative qRT-PCR preliminarily verified the abundance difference of Alistipes in CRC model mice and normal control mice.
[0070] 1. Experimental materials
[0071] The bacterial genomes extracted from feces samples of CRC model mice and normal control mice were verified for the first time by qRT-PCR on the difference in the abundance of Alistipes.
[0072] 2. Test method
[0073] Genome extraction of fecal bacteria: MinkaGene Stool DNA kit (Catalog No. DX1050-02) was used to extract the genome of the stool sample, and a DNA sample with a final concentration of 500-2000 μg / μL was obtained.
[0074] Amplification of the target gene but: real-time fluorescent quantitative PCR was performed using the ChamQ SYBR qPCR Master Mix kit (product number Q331-02 / 03) of Vazyme Company.
[0075] The first set of primers was used for amplification, the upstream primer (Primer1) 5'-CAGCGTCTACATTCAGGGCA-3', the downstream primer (Primer2) 5'-CCGGAGTAGAGCGTG...
Embodiment 2
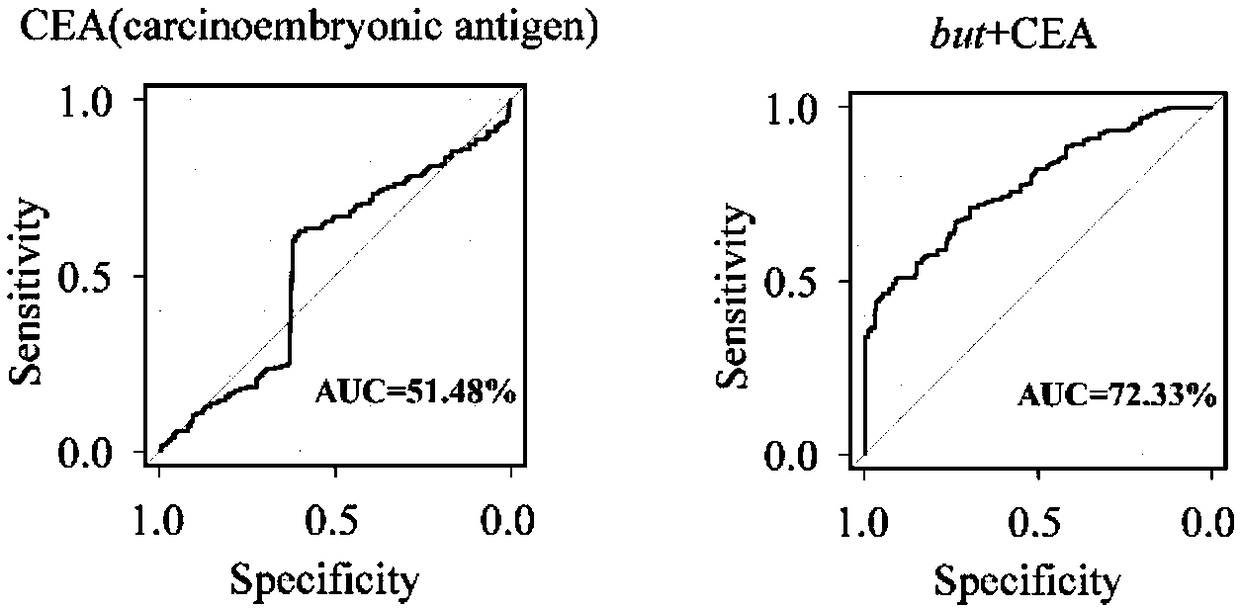
[0082] A ROC curve was constructed to verify the ability of the but gene sequence SEQ ID NO.1 for auxiliary diagnosis to distinguish CRC patients from healthy volunteers.
[0083] The receiver operating curve (ROC) method was used for verification, and the expression level of the signature gene but of characteristic bacteria in the feces of CRC patients and healthy volunteers was used to judge its ability to predict the risk of CRC disease. The result is as figure 2 As shown, it shows that the area under the ROC curve (AUC) for distinguishing CRC patients by basic indicators alone is only 0.51, and after adding the expression indicator of but, the AUC increases to 0.72, which has clinical significance for prognosis.
Embodiment 3
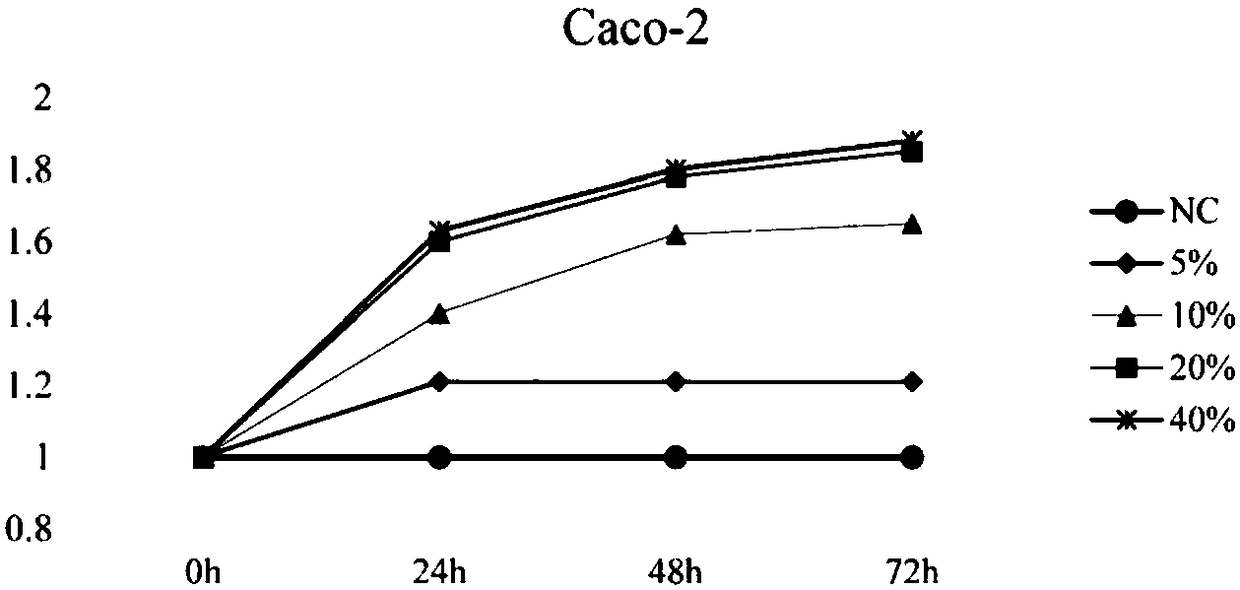
[0085] Used to explore the effect of Alistipes putredinis bacteria on the proliferation of CRC cells.
[0086] Use standard medium (ATCC medium 260) to cultivate Alistipes putredinis bacterial culture value bacterial liquid density OD 600 = 0.5 to confirm that the bacteria have reached log phase. The medium was collected by centrifugation (1000 xg, 15 minutes) and filtered through a 0.22 μm pore size filter to adjust the pH to 7.2-7.4. Then the bacterial culture solution is mixed with the culture medium to a final concentration of 5%, 10%, 20%, 40% of the total culture medium. Different concentrations of bacterial culture solution and colon cancer Caco-2 cells were placed in a 96-well plate (the cell density was 5×10 3 / well) were incubated together for 24h, 48h, and 72h to observe the proliferation of Caco-2 cells. Finally, 10 μL of CCK-8 solution was added to each well, and the plate was incubated at 37° C. for 2 h, and the cell viability was determined by absorbance read...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com