A method for detecting and identifying forest onion snails by fluorescent quantitative PCR
A fluorescence quantitative and forest technology, applied in the field of molecular biology detection and identification, can solve problems such as fluorescence quantitative PCR and other problems that have not yet been seen, and achieve the effect of accurate and convenient result judgment, high sensitivity and strong specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
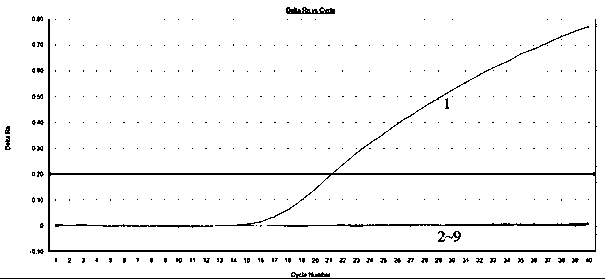
[0017] Embodiment 1: forest onion snail fluorescent quantitative PCR primer specificity test
[0018] 1. Preparation of materials
[0019] The snails for testing are as follows: forest green onion snail, pizza tea snail, bright big snail, worm Yin's snail, cover big snail, Jiangxiba snail, garden green onion snail, scattered big snail.
[0020] The above-mentioned snails were intercepted in the national port quarantine or obtained through domestic investigation and collection. After being confirmed by the National Mollusc Quarantine and Identification Key Laboratory of the former General Administration of Quality Supervision, Inspection and Quarantine, they were stored at -20°C for later use.
[0021] 2. Establishment of fluorescent quantitative PCR method
[0022] 2.1. Design and synthesis of primers: Based on the ITS2 gene sequence of the forest onion snail, the primer design soft Primer Express3 was used to assist in the design and analysis of primers. After the specificit...
Embodiment 2
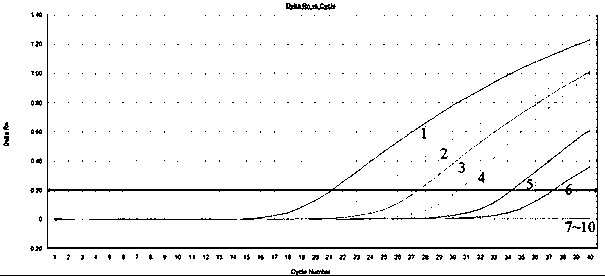
[0030] Example 2: Sensitivity test of forest onion snail fluorescence quantitative PCR detection
[0031]The forest onion snail DNA stock solution (100 ng / μL) extracted in Example 1 was diluted into 10 ng / μL, 1 ng / μL, 100 pg / μL, 10 pg / μL, 1 pg / μL, 100 There are 8 different concentration gradients of fg / μL, 10 fg / μL and 1fg / μL, marked as 1-9 respectively, and curve 10 is the blank control.
[0032] Fluorescent quantitative PCR amplification reaction system: the total volume is 20 μL, including 10 μL of 2×TaqMan PCR Master Mix, 1 μL of upstream and downstream primers and fluorescent probes, 2 μL of 100ng / μL DNA template, 0.5 μL of ROXII, and the rest with sterilized ddH 2 O make up. After mixing, put it into a fluorescent quantitative PCR amplification instrument for amplification.
[0033] The two-step amplification reaction program of fluorescent quantitative PCR was: pre-denaturation at 95°C for 3 minutes; 40 cycles of 10s at 95°C and 40s at 60°C; and the end of the reactio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com