Method for identifying lymphocyte subpopulations infected by EB (epstein-barr) virus and proportion of injected cells in lymphocyte subpopulations and application of method
A technology for lymphocytes and infected cells, which is applied in the field of medical testing, can solve the problems of inability to effectively distinguish the type of lymphocytes infected by EBV and the proportion of infected cells, and achieve improved prognosis, strong universality, and accurate detection results Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] The pretreatment of embodiment 1 lymphocyte
[0060] 1. Extraction of human peripheral blood mononuclear cells (HPBMC)
[0061] 1. Put 6mL of fasting venous blood into a sterile test tube anticoagulated with EDTA, dilute it by 1 time with PBS, and then separate PBMC with lymphocyte separation medium (2000 rpm for 20 minutes);
[0062] 2. Aspirate the middle buffy coat layer, add 10mL PBS, 1400rpm, 10min to wash and precipitate PBMC;
[0063] 3. Discard the supernatant, add 2mL erythrocyte lysate (Solebo erythrocyte lysate product number: Cat#R1010) to lyse, mix well, and incubate at room temperature for 5 minutes;
[0064] 4. After fully mixing, absorb 20 μL of cells and dilute them with 2% glacial acetic acid at 1:3, take the first reading with a cell counter, and record the readings;
[0065] 5. Fill up with PBS, centrifuge at 1400rpm for 10min, discard the supernatant, and obtain human peripheral blood mononuclear cells.
[0066] 2. Surface staining of human perip...
Embodiment 2
[0082] Embodiment 2 probe hybridization and on-machine detection
[0083] 1. Target-specific probe hybridization
[0084] 1. Prepare probe diluent (mixed probes with nucleotide sequences as shown in SEQ ID NO.1, SEQ ID NO.2 and SEQ ID NO.3). Preheat the Target Probe Diluent to 40°C in advance, and dilute the 20× probe to 1×. Each sample requires 100 μL of Probe Diluent. Add 5 μL of EBER probe to 95 μL of Target Probe Diluent, and mix well by pipetting up and down;
[0085] 2. Add 100 μL of the diluted probe directly into the cell suspension, and be careful not to hit the probe on the tube wall. Vortex evenly, incubate at 40°C for 2 hours, mix the sample upside down after 1 hour of hybridization, and continue to complete the hybridization for the last 1 hour;
[0086] 3. Add 1 mL of Wash Buffer to each sample, invert to mix, centrifuge at 800×g for 5 min, discard the supernatant until 100 μL remains, and use the residual liquid to resuspend the cells;
[0087] 4. Prepare W...
Embodiment 3
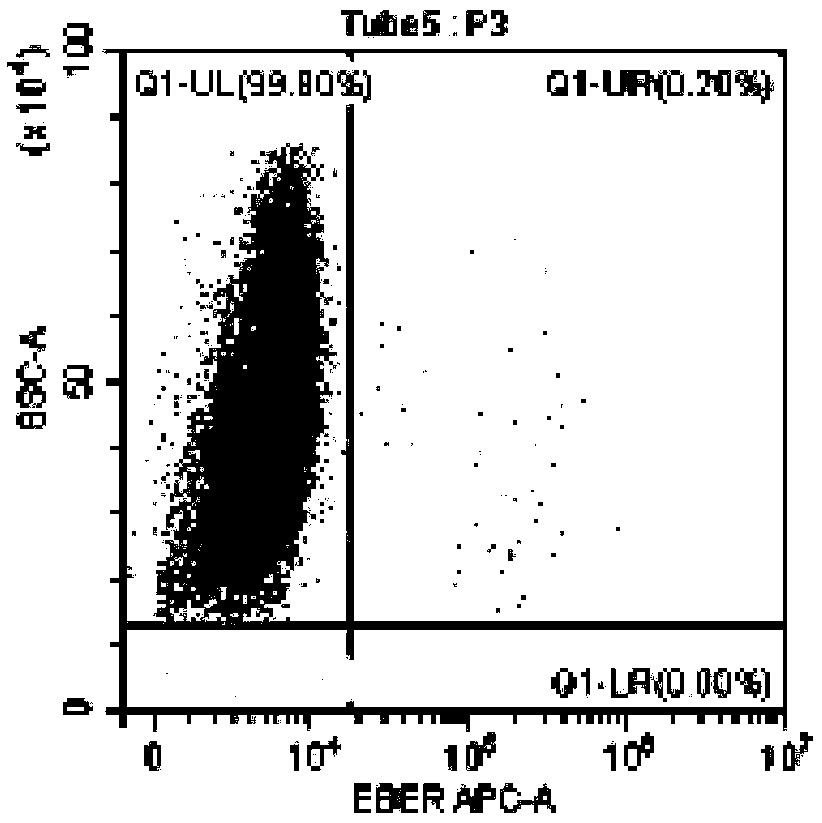
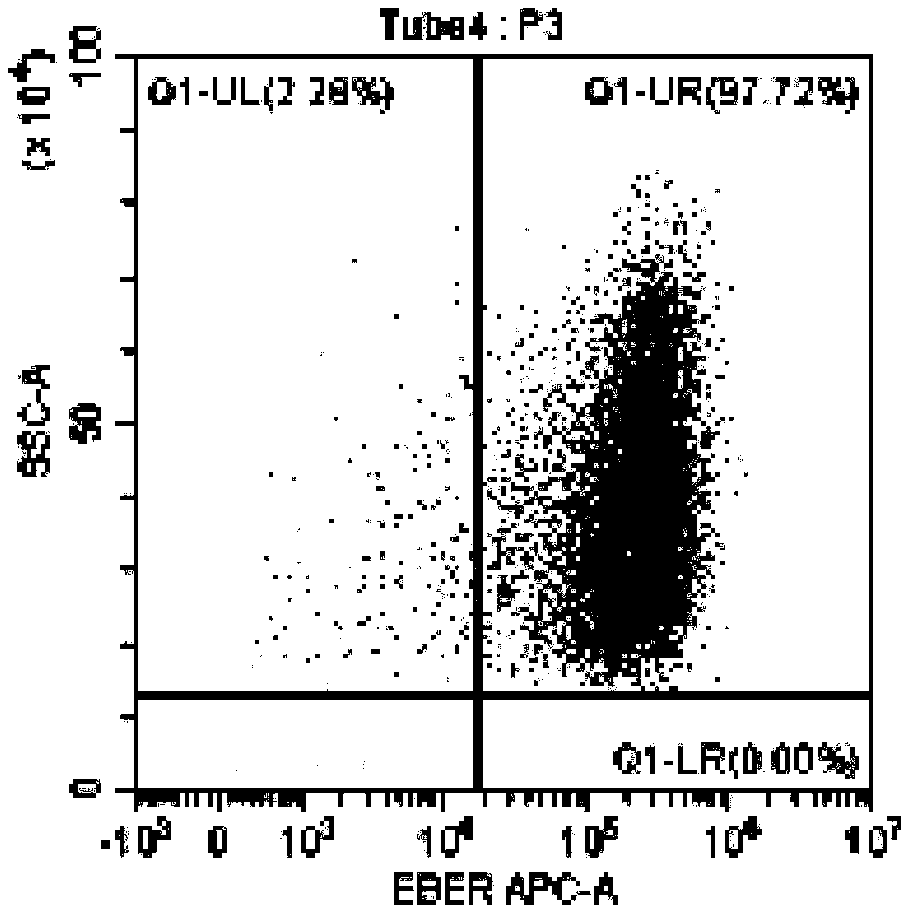
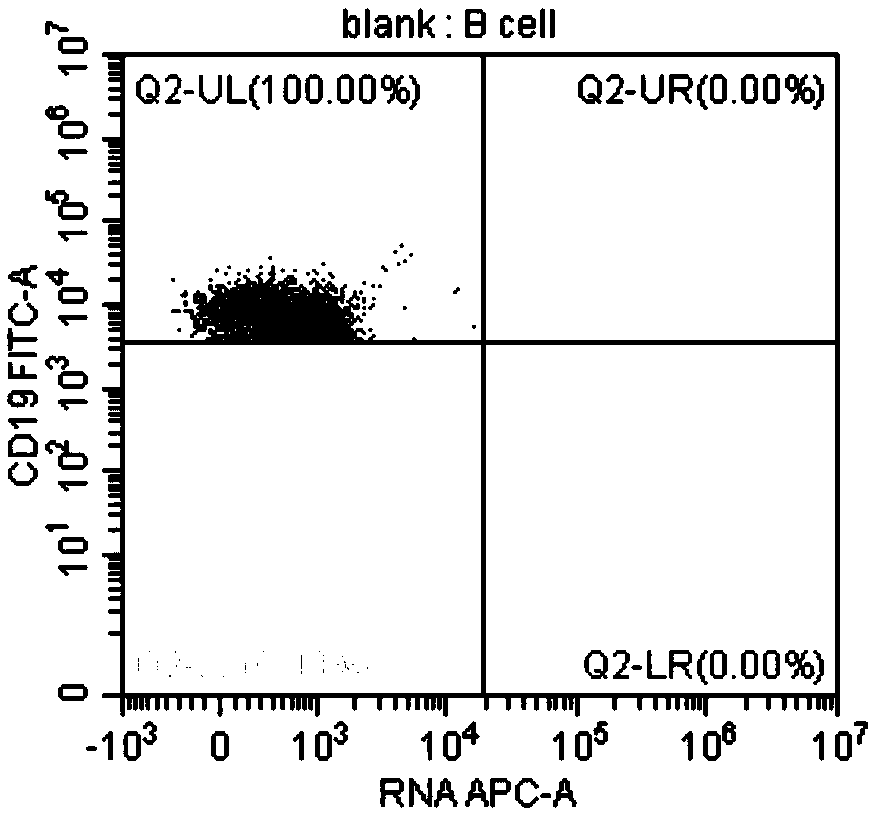
[0102] In this example, the method for identifying EBV-infected lymphocyte subgroups and the proportion of infected cells in the subgroup provided by the present invention was used to detect the EBV infection of peripheral blood lymphocytes of an EBV-positive patient. Wherein, the method provided in Example 1 was used for pretreatment of lymphocytes, and the method provided in Example 2 was used for probe hybridization and on-machine detection. The objects tested on the machine are CD3+T, CD4+T, CD8+T, CD19+B, CD56+NK EBV infection.
[0103] The test results are shown in the figure.
[0104] in, Figure 3A It is the detection result chart of the blank control tube of CD19+ B cells without adding EBER probe in this patient, Figure 3B Diagram of the detection results of CD19+ B cells added with EBER probes for this patient; from Figure 3A and Figure 3B As can be seen in , the results show that 11.85% of the CD19+ B cells in this patient were infected with EBV. Figure 4A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com