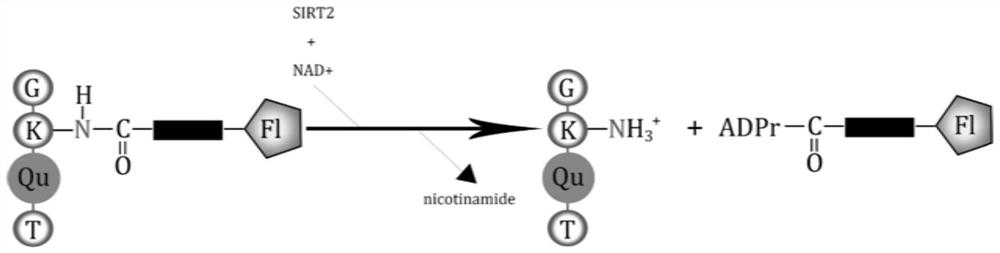
A kind of fluorescent probe and its application in detecting sirt2 enzyme activity
A fluorescent probe and probe technology, applied in fluorescence/phosphorescence, measurement device, material analysis by optical means, etc., to achieve the effect of low cost, simple operation and reduced error
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
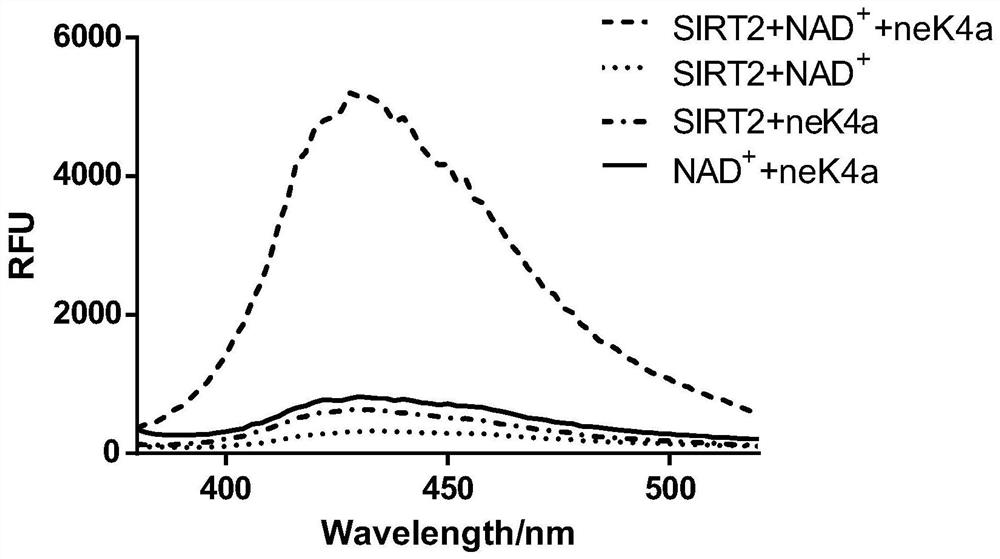
[0036] Verify that fluorescent probes are not affected by SIRT2 or NAD + The effect of acting alone, including the following steps:
[0037] (1) Configure the following four groups of samples
[0038] Sample 1: Add 1 μl NAD to 47 μl buffer solution (50 mM HEPES; 100 mM KCl; 0.001% Tween-20; 0.05 mg / ml BSA; 200 mM TCEP; pH: 7.4, the same below) + (50mM, the same below), 1μl fluorescent probe (1mM, the same below), and finally add 1ul SIRT2 (15μM, the same below);
[0039] Sample 2: Add 1 μl fluorescent probe to 48 μl buffer, and finally add 1ul sirt2;
[0040] Sample 3: Add 1 μl NAD to 48 μl buffer + , and finally add 1ul of sirt2;
[0041] Sample 4: Add 1 μl fluorescent probe and 1 μl NAD to 48 μl buffer + ;
[0042] (2) The four groups of samples were all reacted at 37° C. for one hour, and after the reaction, the fluorescence spectrum from 380 nM to 580 nM was detected with a Flex Station3 multifunctional microplate reader under excitation of 320 nM.
[0043] Depend o...
Embodiment 2
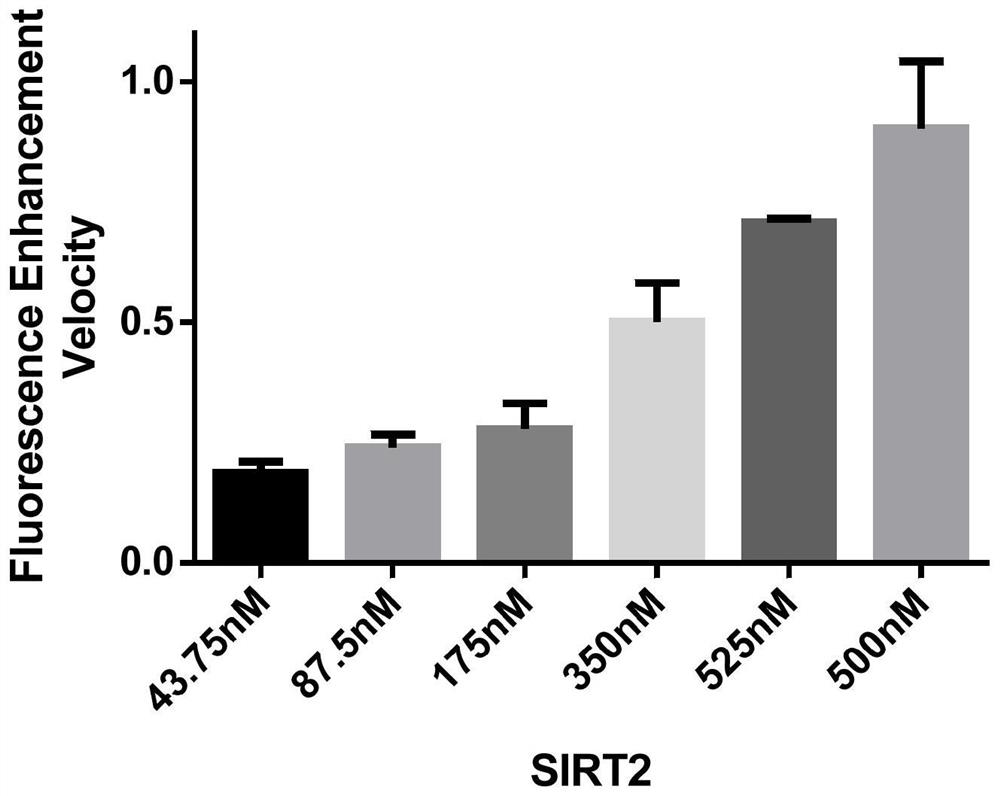
[0045] The detection of the sensitivity of the fluorescent probe to the SIRT2 enzyme includes the following steps:
[0046] Add 1 μl fluorescent probe (1 mM) and 1 μl NAD to 47 μl buffer + (50mM), and finally add different concentrations of 1μl SIRT2 (final concentrations are 43.75nM, 87.5nM, 175nM, 350nM, 425nM, 500nM), using FlexStation3 multifunctional microplate reader at 37°C, excitation wavelength 320nM, emission wavelength 416nM continuous Detect for 30 minutes, and count the rate of fluorescence change within 30 minutes.
[0047] Depend on image 3 It can be seen that under the condition that the concentration of the fluorescent substrate remains unchanged, as the concentration of SIRT2 protein increases, the fluorescence enhancement rate of the system also increases.
Embodiment 3
[0049] The detection of the specificity of the fluorescent probe to the SIRT2 enzyme includes the following steps:
[0050] (1) Configure the following three groups of samples
[0051] Sample 1: Add 1 μl SIRT1 (15 μM), 1 μl NAD+, 1 μl fluorescent probe to 47 μl buffer.
[0052] Sample 2: Add 1 μl SIRT2 (15 μM), 1 μl NAD+, 1 μl fluorescent probe to 47 μl buffer.
[0053] Sample 3: Add 1 μl SIRT3 (15 μM), 1 μl NAD+, 1 μl fluorescent probe to 47 μl buffer.
[0054] (2) The three groups of samples were all reacted at 37°C, and the Flex Station3 multifunctional microplate reader was used to continuously detect the excitation wavelength of 320nM and the emission wavelength of 416nM for 30min, and the fluorescence change rate within 30min was counted.
[0055] Depend on Figure 4 It can be seen that at the same concentration, the sample group added with SIRT1 protein and SIRT3 protein basically has no fluorescence signal, while the SIRT2 sample group has a significant increase in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com