Few-sample whole genome DNA (deoxyribonucleic acid) methylation detection method and kit
A whole genome and detection method technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of not having universal applicability, high cost, inapplicability, etc., to ensure the efficiency of joint connection and improve efficiency , strong practical effect
Inactive Publication Date: 2018-11-13
SHANGHAI JIAO TONG UNIV
View PDF3 Cites 5 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0004] There are two main disadvantages of the existing technology: ①The minimum required sample input amount is 50ng DNA or 75,000 cells, which is still not suitable for MeDIP-seq analysis of a small amount of source samples; ②It needs to use commercial kits and fully automatic Sample processing system for MeDIP-seq experiments is expensive and not universally applicable
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
[0037] The following describes the preferred embodiments of the present invention with reference to the accompanying drawings to make the technical content clearer and easier to understand. The present invention can be embodied in many different forms of embodiments, and the protection scope of the present invention is not limited to the embodiments mentioned herein.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
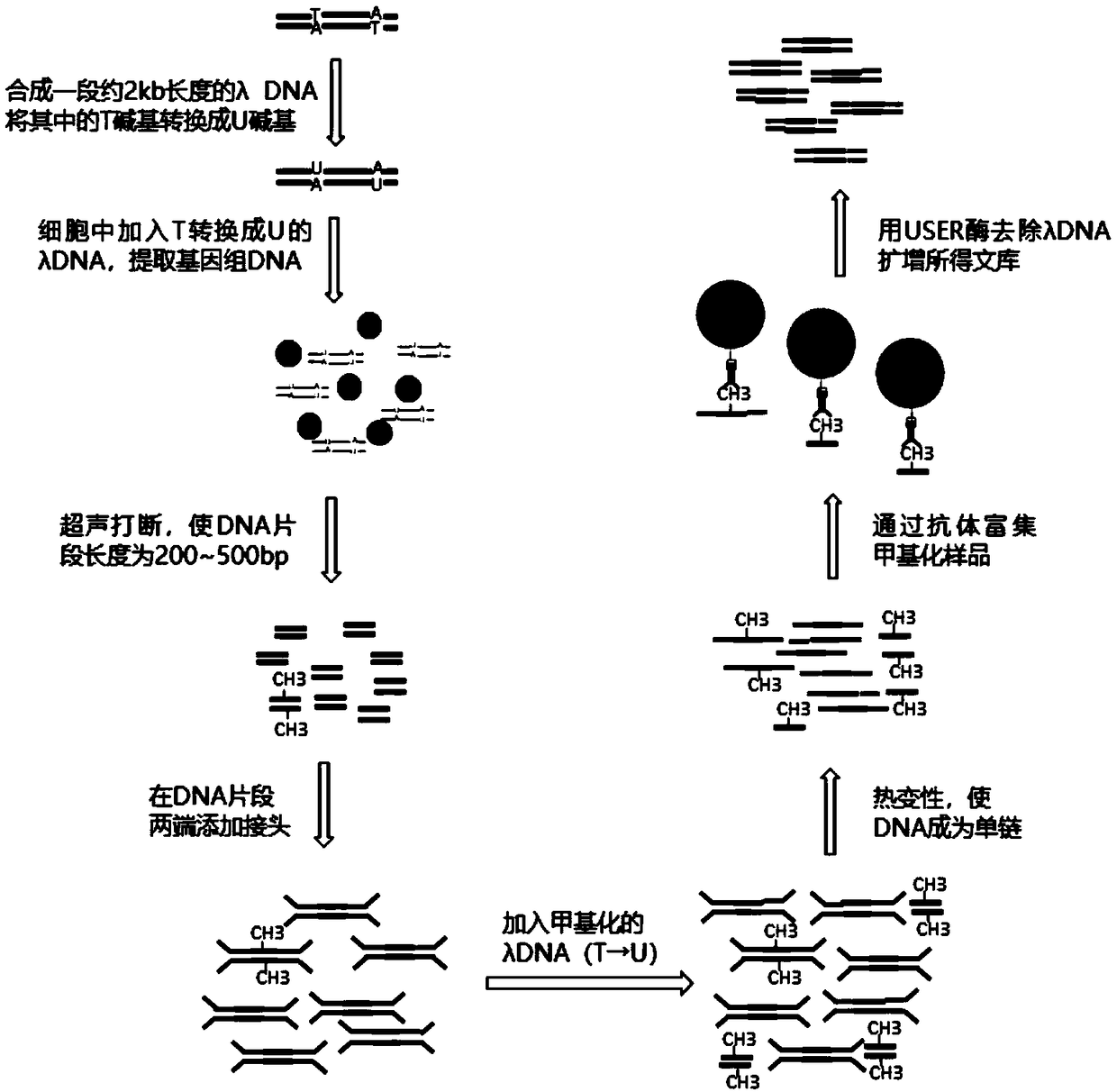
The invention discloses a few-sample whole genome DNA (deoxyribonucleic acid) methylation detection method, and relates to a method for detecting less-sample (300pg DNA or 50 cells) whole genome DNA methylation by an improved MeDIP-Seq technique based on high-throughput sequencing. According to the method, lambda DNA containing dUTP is added into a small number of initial samples to reduce sampleloss, base-building and immuno-precipitation efficiency is improved, MeDIP-DNA is digested by USER enzyme, so that added methylation lambda DNA is removed, and a methylation sample sequencing libraryto be detected cannot be polluted by the methylation lambda DNA. According to the method, needed sample amount is 300pg DNA or 50 cells, the method is more applicable to MeDIP-Seq analysis of a smallnumber of source samples, operation method of a MeDIP experimental technique and a kit matched with the experimental technique is simple and convenient, specific commercial kits and experimental devices are omitted, cost is low, and the method has wide applicability and universality.
Description
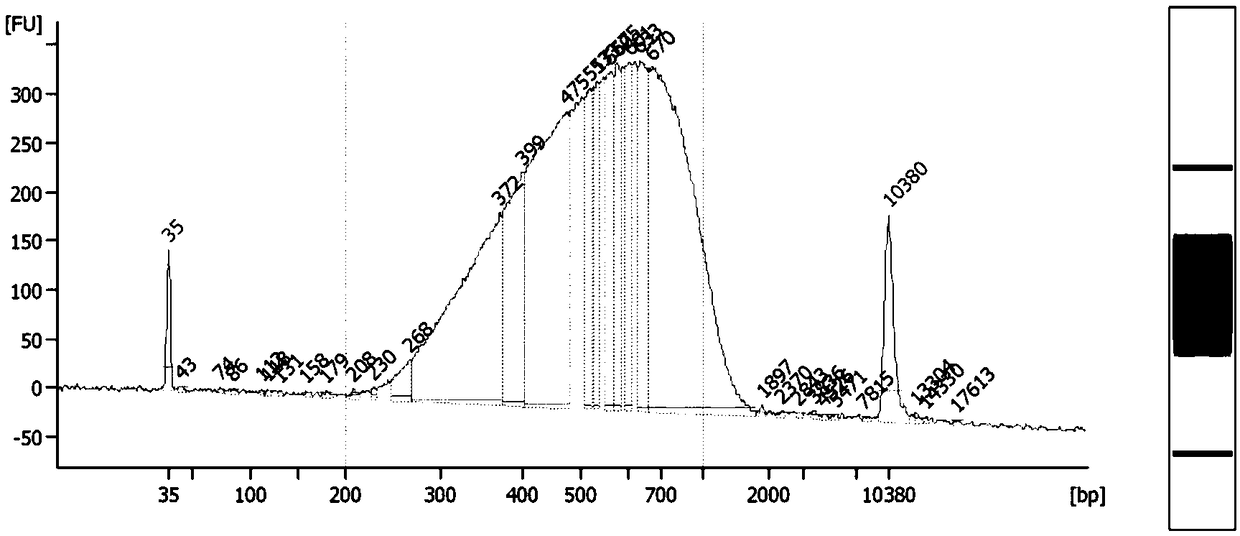
technical field [0001] The present invention relates to a method for detecting DNA methylation in the whole genome, in particular to a method for detecting whole genome DNA in a small amount of samples (300pg DNA or 50 cells) by improving MeDIP-Seq (Methylated DNA immunoprecipitation sequencing) technology based on high-throughput sequencing method of methylation. Background technique [0002] At present, the minimum amount of DNA required to detect the DNA methylation of the whole genome of cells by MeDIP-seq method is 50ng. If cells are used as the starting sample, at least ~75000 cells are required (Taiwo O et al., Nat Protoc, 2012), The specific experimental process of this method is: extraction of cellular genomic DNA → ultrasonic fragmentation of genomic DNA and detection of ultrasonic fragment size by Agilent Bioanalyzer 2100 → end repair of ultrasonic fragments, addition of dA at the 3' end, adapter ligation, and purification and quantification of the ligated product...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/6869C12Q1/6806
CPCC12Q1/6806C12Q1/6869C12Q2535/122C12Q2523/301C12Q2525/191C12Q2531/113C12Q2521/531
Inventor 康亚妮赵小东邵志峰胡丛霞
Owner SHANGHAI JIAO TONG UNIV
Features
- Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com