Reagent kit and method for detecting DNA (deoxyribonucleic acid) methylation and application
A methylation and kit technology, applied in the field of molecular biology and DNA methylation detection, can solve the problem of methylation analysis that is not applicable to a small number of source samples, cannot be used for genome-wide methylation research, and has no universal applicability and other issues, to achieve the effects of small sequencing scale, lower sequencing cost, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
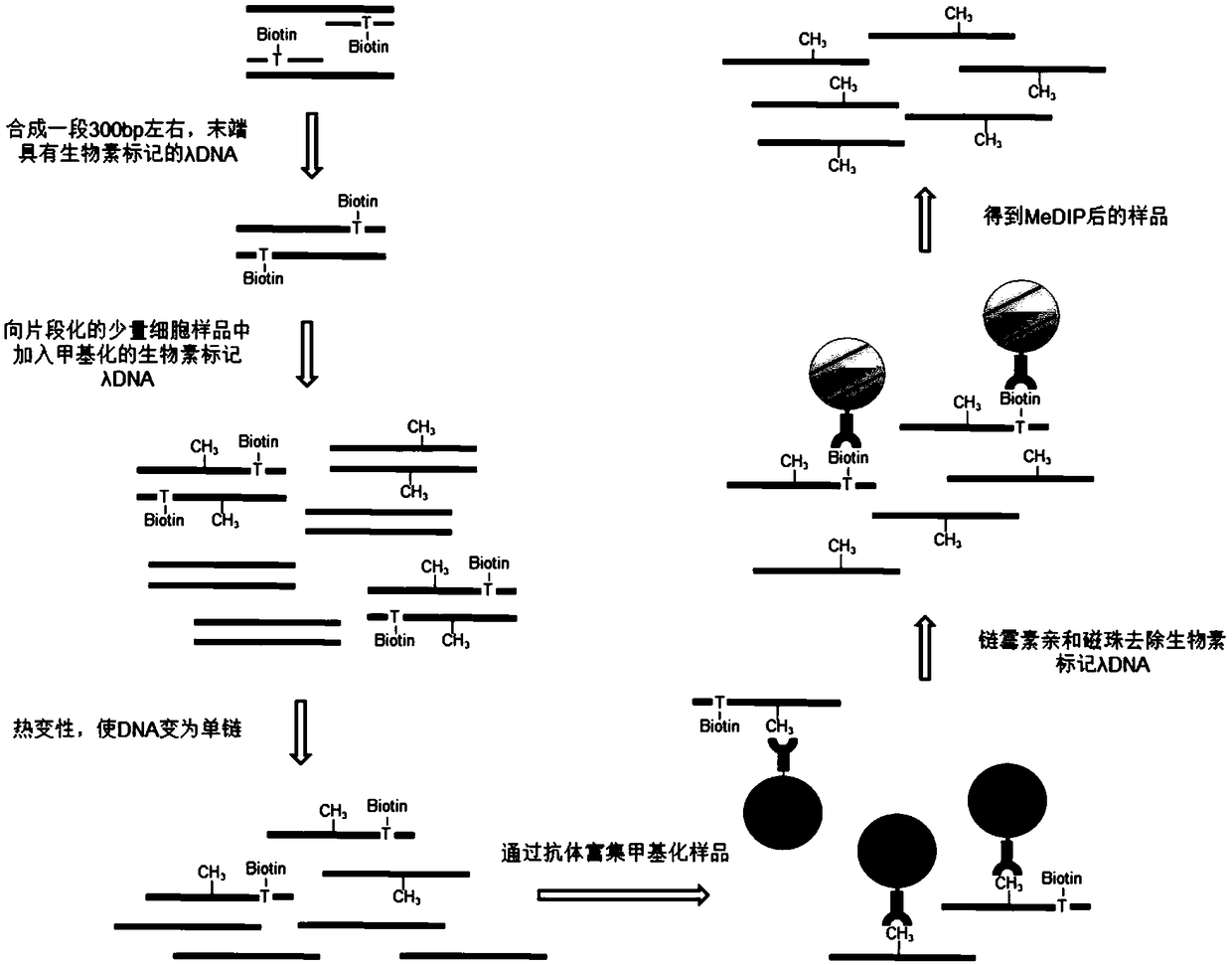
[0039] Below in conjunction with specific embodiment, further illustrate the present invention. It should be understood that these examples are only used to illustrate the present invention and are not intended to limit the scope of the present invention. The experimental materials and reagents used in the following examples can be obtained from commercially available channels unless otherwise specified.
[0040] Preparation of biotinylated λ-DNA
[0041]1. Prepare biotin-modified λ-DNA (Bio-λ-DNA) with a fragment length of about 300 bp by PCR amplification. The PCR reaction system is as follows:
[0042] λ-DNA
60ng
Taq PCR Master Mix (BBI, B639295)
25ul
Upstream primer (SEQ No.1)
0.4uM
Downstream primer (SEQ No.2)
0.4uM
double distilled water
to 50ul
[0043] PCR reaction conditions:
[0044]
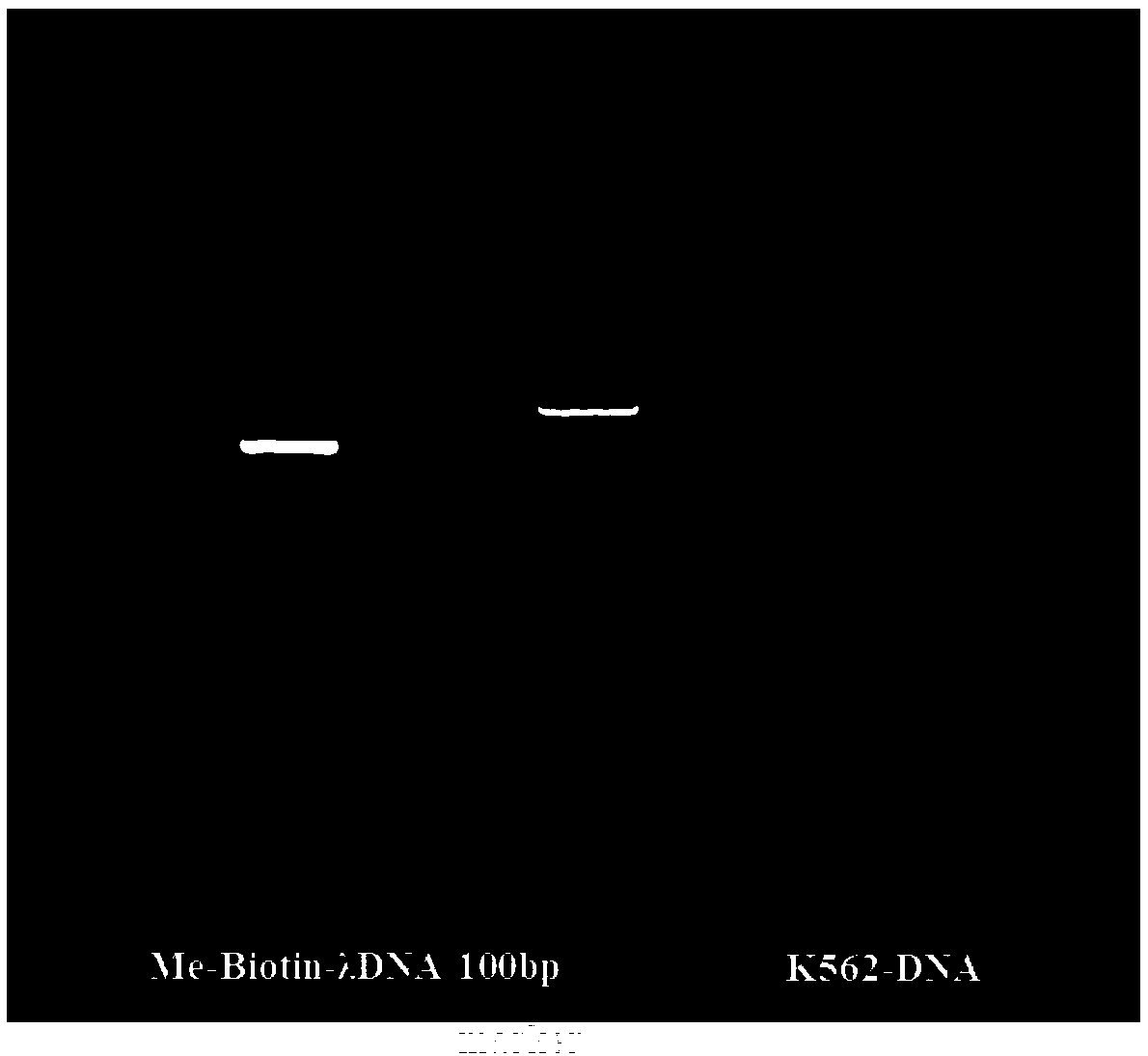
[0045] 2. Use AxyPrep TM The PCR purification kit (Axygen, AP-PCR-250G) was used to purify the PCR product and quan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com