Corn chloroplast genome SSR site and application in variety identification
A genome and corn leaf technology, applied in the field of crop molecular biology, can solve problems such as unrecorded SSR polymorphic sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
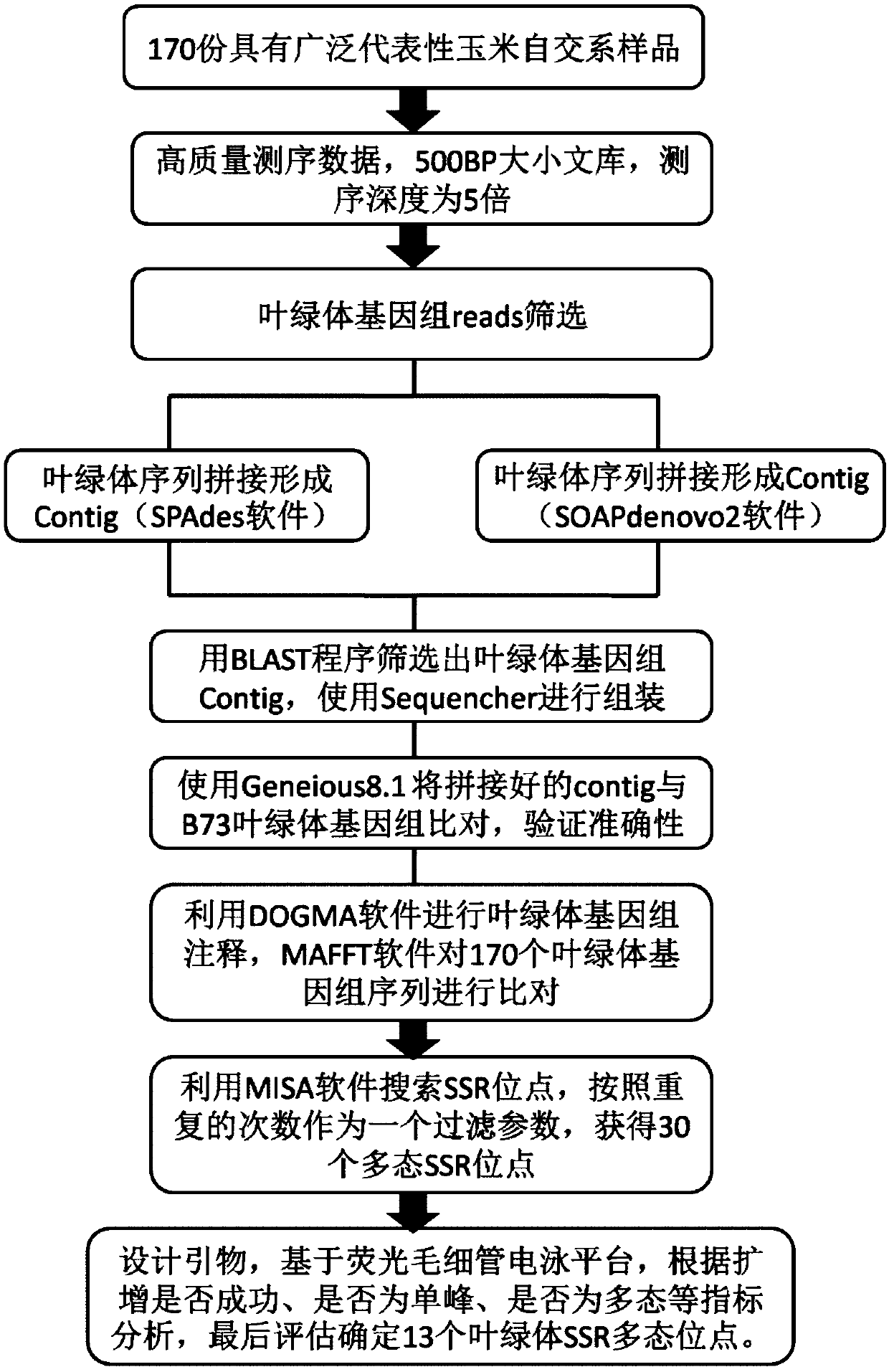
[0030] Example 1 Using the combination of SSR polymorphic sites provided by the present invention to construct a corn variety chloroplast DNA-SSR fingerprint database
[0031] DNA extraction of corn varieties: Each sample was seeded with seeds, given light, and formed green shoots. The method of DNA extraction was mixed plant DNA extraction, and the green leaves of 30 individual plants were mixed. The specific steps of DNA extraction were carried out according to the molecular identification standard of maize DNA (Wang Fengge et al., 2014). Dilute the DNA to form a working solution with a concentration of 20ng / μL.
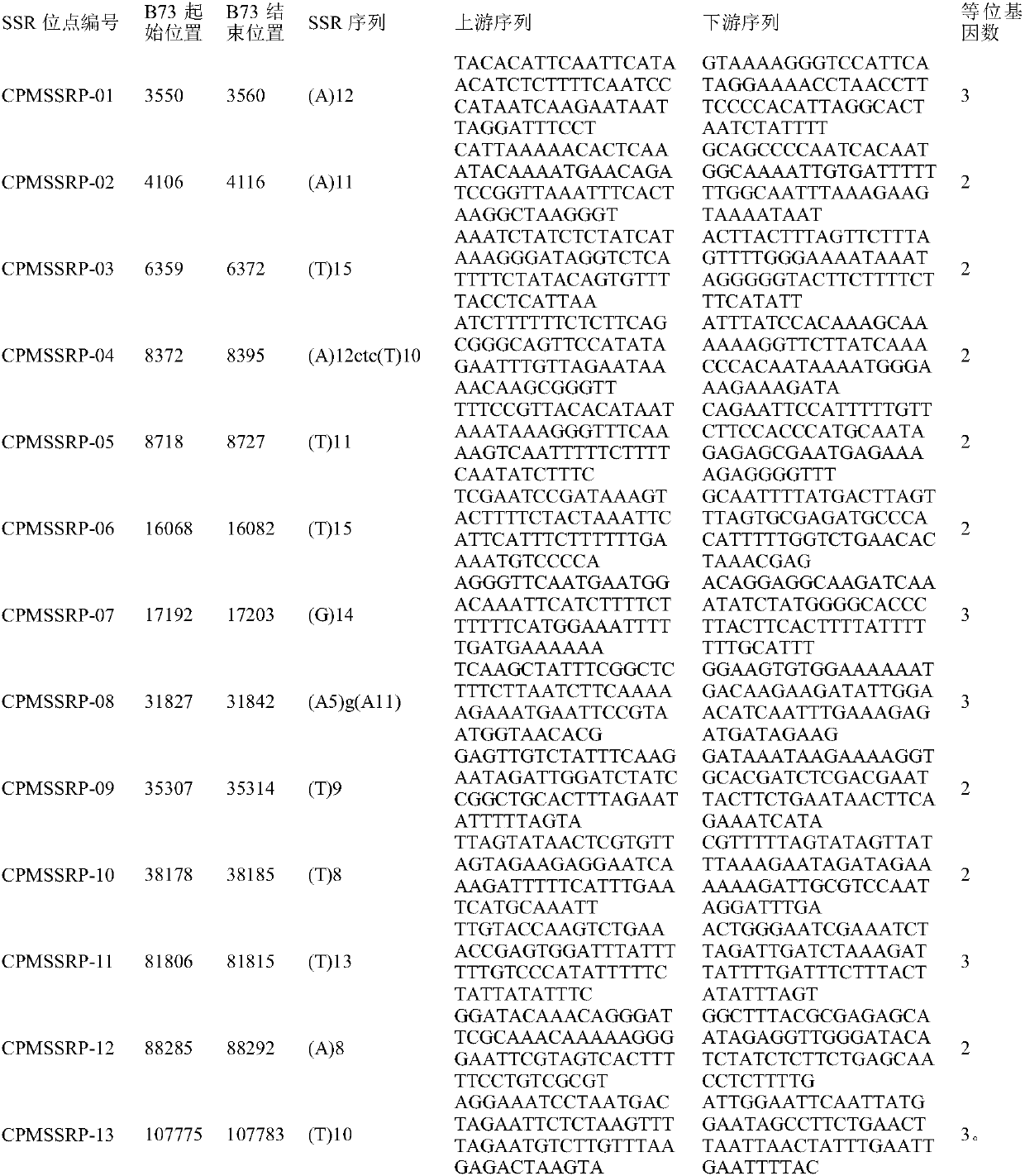
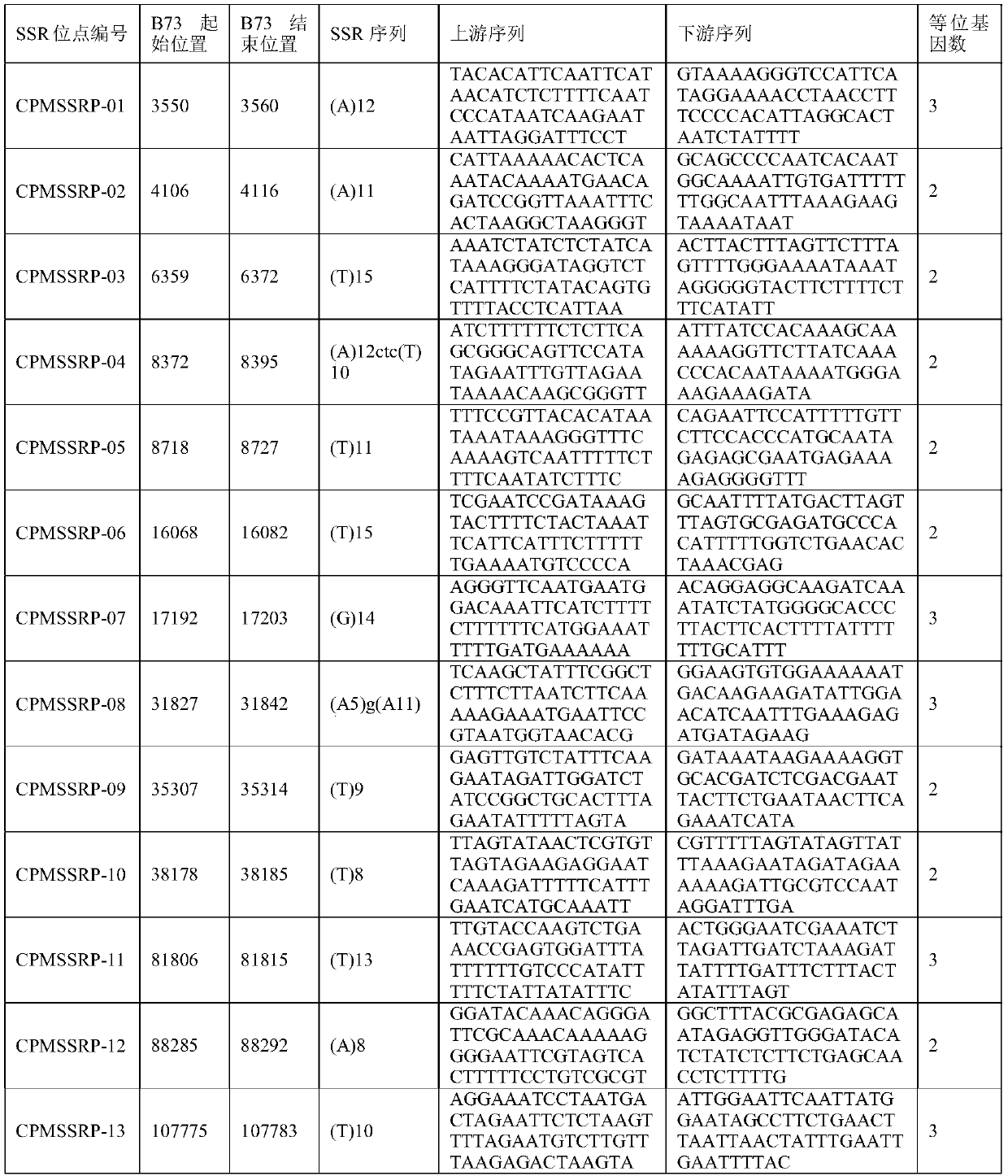
[0032] Design and synthesis of primers: The 13 SSR sites (Table 1) provided by the present invention are single base length polymorphisms, so primers can be designed for this set of sites based on the fluorescent capillary electrophoresis platform. A pair of primers was designed for each site based on its flanking sequence, and the 5' end of one primer was labeled...
Embodiment 2
[0035] Example 2 Using the maize chloroplast genome SSR polymorphic sites provided by the present invention to perform maternal traceability analysis
[0036] For the maize hybrid A to be identified, DNA is extracted from the green leaves formed by seedlings, and the 13 SSR sites provided in the present invention are used for PCR amplification and fluorescent capillary electrophoresis to obtain fingerprint data. Concrete method is with embodiment 1. Based on the comparison of the SSR fingerprint data of sample A (Jingke 968) with the chloroplast SSR marker fingerprint database of known maize inbred varieties (refer to the external platform of the Chinese maize standard DNA fingerprint database, the website is: www.maizedna.org), determine The chloroplast fingerprint information of the sample to be tested is the same as that of the B inbred line (Jing 724), and it is speculated that the female parent of the hybrid A (Jingke 968) is B (Jing 724).
Embodiment 3
[0037] Example 3 Using the SSR polymorphic sites of the maize chloroplast genome provided by the present invention to identify the reciprocal cross of maize samples
[0038] For the reciprocal hybrid sample C (Zhengdan 958) to be identified, the DNA was extracted from the green leaves of the seeds. Based on the DNA fingerprint data of the parental parents of Zhengdan 958 in the chloroplast SSR marker fingerprint database of known maize inbred varieties, the polymorphic positions in the parental parents of the sample Zhengdan 958 were selected from 13 polymorphic SSR sites. Point PCR amplification, after fluorescence electrophoresis data analysis, obtain the fingerprint of sample C, the specific method is the same as embodiment 1. The fingerprint data of sample C to be tested was compared with the fingerprint data of its female parent (sample D, Zheng 58) and male parent (sample E, Chang 7-2). If the fingerprint data of sample C is the same as that of the female parent of samp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com