Primer pair and kit for detecting P53 gene epigenetic modification difference in peripheral blood free DNA
An epigenetics, peripheral blood technology, applied in recombinant DNA technology, DNA/RNA fragment, determination/inspection of microorganisms, etc., can solve the problems of unclear epigenetic modification function and little attention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Primer pair set for detecting the difference in epigenetic modification of P53 gene in peripheral blood cell-free DNA
[0048] According to the sequences of different segments of the P53 gene, PCR primers were designed and a large number of screening experiments were carried out, and finally a set of primer pairs with strong specificity and high sensitivity for detecting the difference in epigenetic modification of the P53 gene in peripheral blood cell-free DNA was obtained. , the primer pair set consists of the nucleotide sequences shown in SEQ ID No.1 to SEQ ID No.10 in the sequence listing;
[0049] Among them, the sequences of SEQ ID No.1 and SEQ ID No.2 are respectively the upstream primer and the downstream primer for amplifying the exon 3 region of the P53 gene with a length of 140bp (P53-E3-140), and the sequence SEQ ID No.3 is A probe for amplifying the fragment of this region; SEQ ID No.4 and SEQ ID No.5 are respectively the upstream primer and the d...
Embodiment 2
[0078] Example 2 Kit for detecting the difference in epigenetic modification of P53 gene in cell-free DNA of peripheral blood
[0079] In peripheral blood cell-free DNA, the kit for detecting the difference in epigenetic modification of P53 gene includes the primers in Example 1 consisting of the nucleotide sequences shown in SEQ ID No. 1 to SEQ ID No. 10 of the Sequence Listing Pair group, PCR reaction solution (Premix Ex Taq TM ), QC control DNA samples and ddH 2 O;
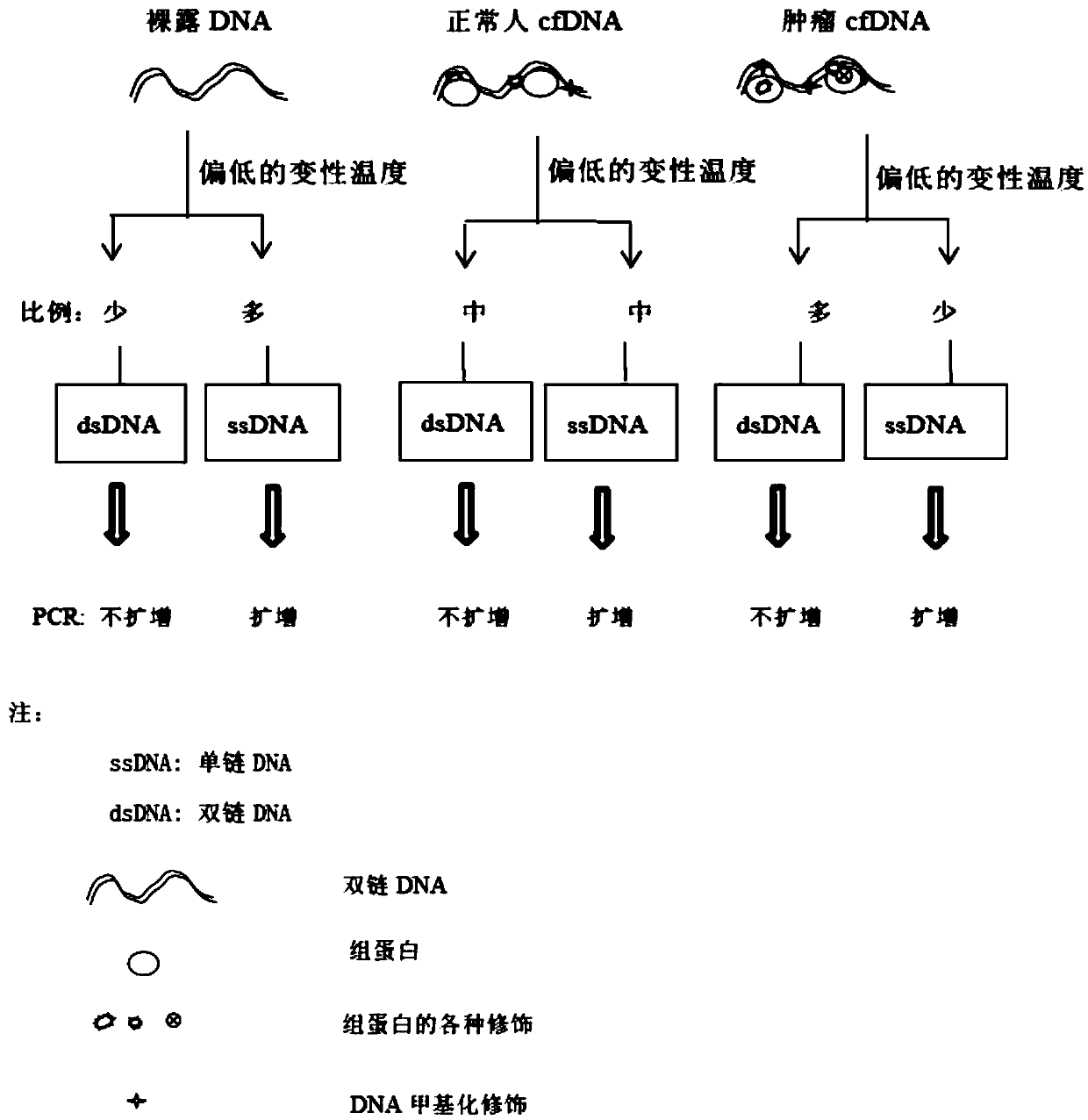
[0080] The quality control control DNA sample is naked DNA, for example, it can be a PCR product without epigenetic modification; further, in order to better simulate the concentration of free DNA, the PCR product needs to be highly diluted to make it equal to the sample concentration. Re-amplification as a template; wherein, the quality control control DNA sample includes a 140bp naked DNA fragment in the exon 3 region of the P53 gene, a 204 bp naked DNA in the P53 gene exon 3 region, and a P53 gene exon 4 r...
Embodiment 3
[0081] Embodiment 3 Detection method and process
[0082] 1) Extract cell-free DNA from peripheral blood;
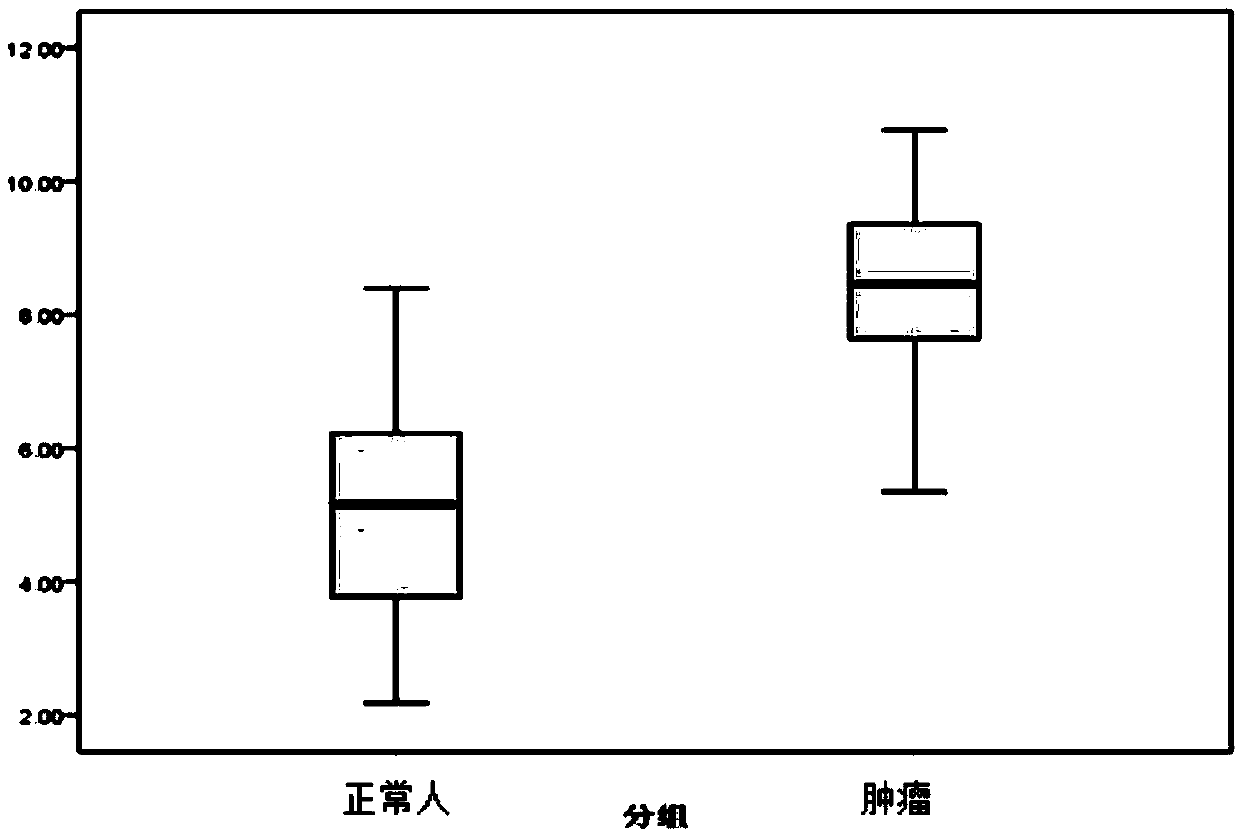
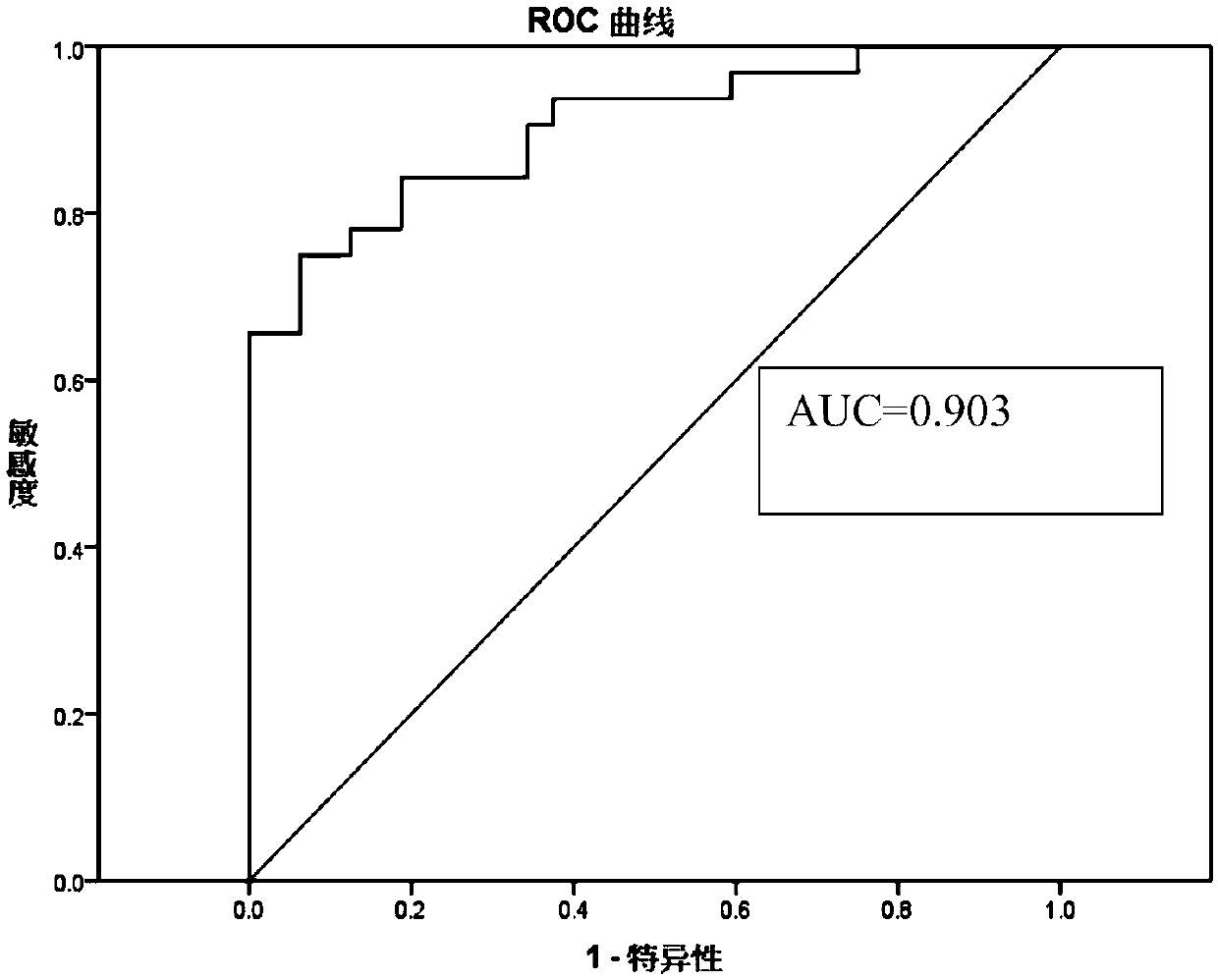
[0083] 2) Using a primer pair set consisting of the nucleotide sequences shown in SEQ ID No. 1 to SEQ ID No. 10 in the sequence table, the peripheral blood free DNA and quality control control DNA samples were denatured at high temperature and low temperature respectively. Real-time fluorescence quantitative PCR amplification under conditions,
[0084] The qPCR reaction system of each primer set of the reaction body is shown in Table 1-4:
[0085] Table 1. x1 sequence qPCR reaction system
[0086]
[0087] Table 2. x2 sequence qPCR reaction system
[0088]
[0089]
[0090] Table 3. x3 sequence qPCR reaction system
[0091]
[0092] Table 4. x4 sequence qPCR reaction system
[0093]
[0094] The reaction conditions are:
[0095] High Temperature (HT): Pre-denaturation at 95℃ for 5min; 94℃ for 5s, 60℃(x1,x3) / 58℃(x2,x4) for 15 seconds, 72℃ for 30s, 50 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com