Peach transcription factor PpERF.A16 gene, protein as well as recombinant expression vector and application thereof
A technology of transcription factors and expression vectors, applied in the fields of application, genetic engineering, plant gene improvement, etc., to achieve the effects of reducing agricultural costs, increasing commodity value, and extending shelf life
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Construction of phylogenetic tree and related gene qRT-PCR analysis
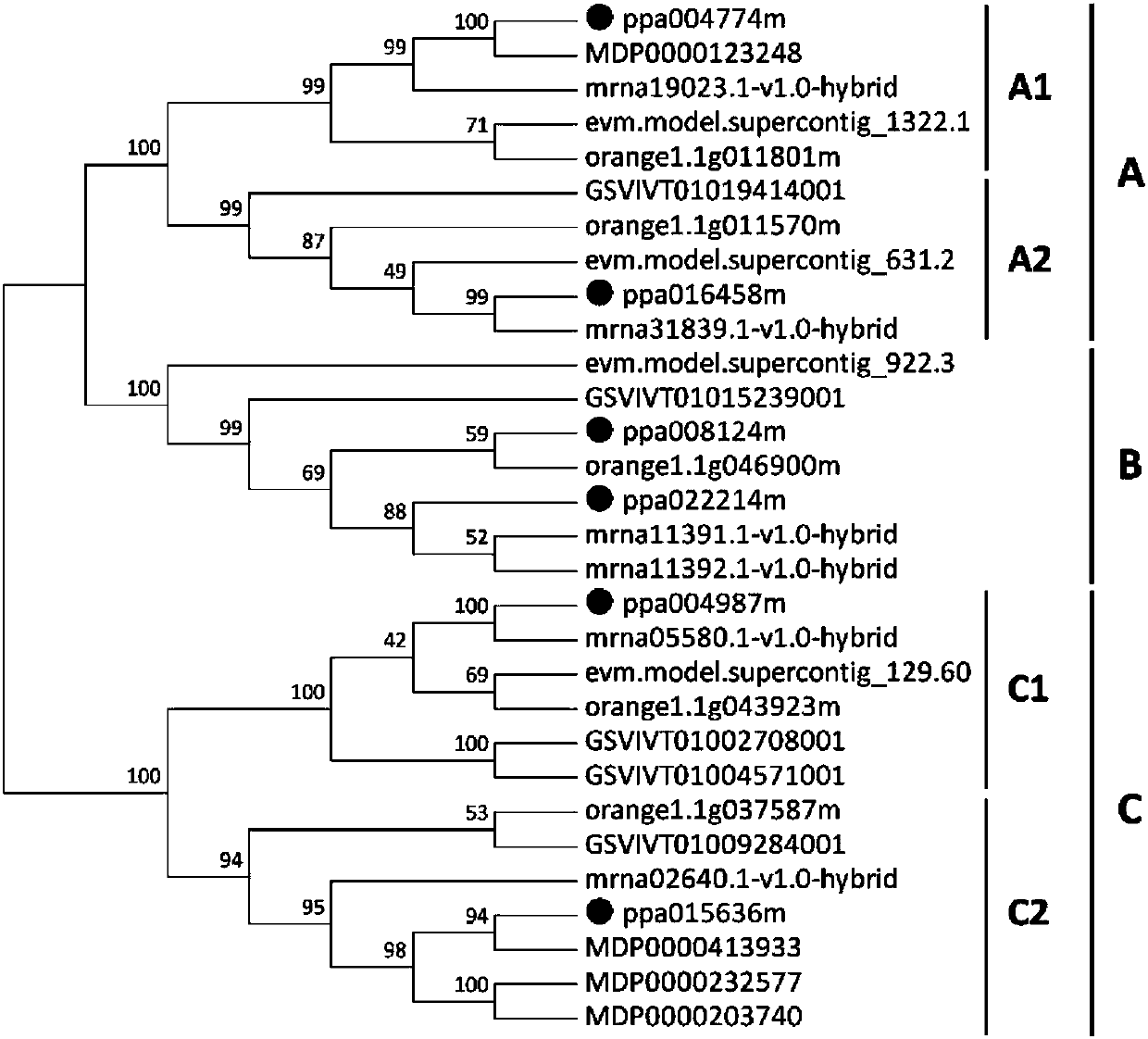
[0044] 1. Download the ACS from the peach genome database (Rosaceae, http: / / www-RuxaA.Org) and other fruit trees (including apple, strawberry, papaya, orange and grape) (http: / / PosiZoM.jig.doe.gov) , ACO and ERF family members of the nucleotide and amino acid sequences. All genes were predicted in the National Center for Bioinformatics (NCBI; HTTPS: / / www.NcB.NLM.NIH.GOV / ), and the predicted genes were classified. The downloaded amino acid sequences were aligned by CulsTAL X, and a neighbor-joining (NJ)-based phylogenetic tree was constructed using the software MEGA 6 . the result shows:
[0045] 1.1 Six ACS gene family members were detected from the peach genome, which can be divided into three groups A, B, and C. Groups A and C consist of two subgroups ( figure 1 ). These genes were named PpACS.A1, PpACS.A2, PpACS.B1, PpACS.B2, PpACS.C1 and PpACS.C2 (Table 1).
[0046] 1.2 36 members o...
Embodiment 2
[0052] Example 2 PpERF.A16 Gene Isolation Cloning and Vector Construction
[0053] 1. Using the plant total RNA extraction kit of polyphenols and polysaccharides (purchased from TIANGEN Company, operated according to the kit instructions) to extract RNA from peach pulp, the first-strand cDNA obtained by reverse transcription was used to amplify Increase the full length of the PpERF.A16 gene. The primer pair for the amplification gene is: PpERF.A16-F1: (SEQ ID NO.3); PpERF.A16-R1: (SEQ ID NO.4). The 50 ul reaction system included 2 ul of the above primers, 17 ul of ddH2O, 25 ul of 2×Buffer, 1 ul of dNTP, 2 ul of cDNA template, and 1 ul of high-fidelity enzyme (Phanta Super-Fidelity DNA Polymerase) (purchased from Vazyme). The PCR reaction was completed on the Eppendor PCR amplification instrument according to the following program: 95°C, pre-denaturation for 3 minutes, denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 1 minute, 30 the...
Embodiment 3
[0055] Example 3 PpERF.A16 gene overexpression and construction of silencing vector
[0056] 1. According to the analysis of the multiple cloning site of the pCAMBIA-1301 vector and the restriction site on the coding region sequence of the PpERF.A16 gene, XbaI and HindIII were selected as endonucleases. According to the general principles of primer design, primers with restriction sites were designed with Primer 5.0 software, and the sequences of the primer pairs are as follows:
[0057] PpERF.A16-F1: ttggatccAGGAATGTGTGGCGGTGCTAT (SEQ ID NO. 3)
[0058] PpERF.A16-R1: aatctagaTTACGGAGCAGAAACGCGGTCG (SEQ ID NO. 4)
[0059] Using the extracted plasmid from the preserved bacterial liquid with correct sequencing as a template, clone the gene containing the restriction endonuclease site. The annealing temperature of PCR amplification is 60° C., and the PCR reaction system and amplification procedure are the same as in Example 1. The pCAMBIA-1301 vector was double-enzyme-digested...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com