Application of sucrose lethal gene SacB in gene deleted reverse screening marker, and marker-free deleted suicide vector of sucrose lethal gene SacB
A suicide carrier and reverse screening technology, which is applied in the direction of introducing foreign genetic material, carrier, nucleic acid carrier, etc., can solve the problems of limited range of resistance gene selection, trace deletion, non-transcription of downstream genes, etc., and achieve biological Universality, effects of avoiding polar effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1, the amplification of SacB gene
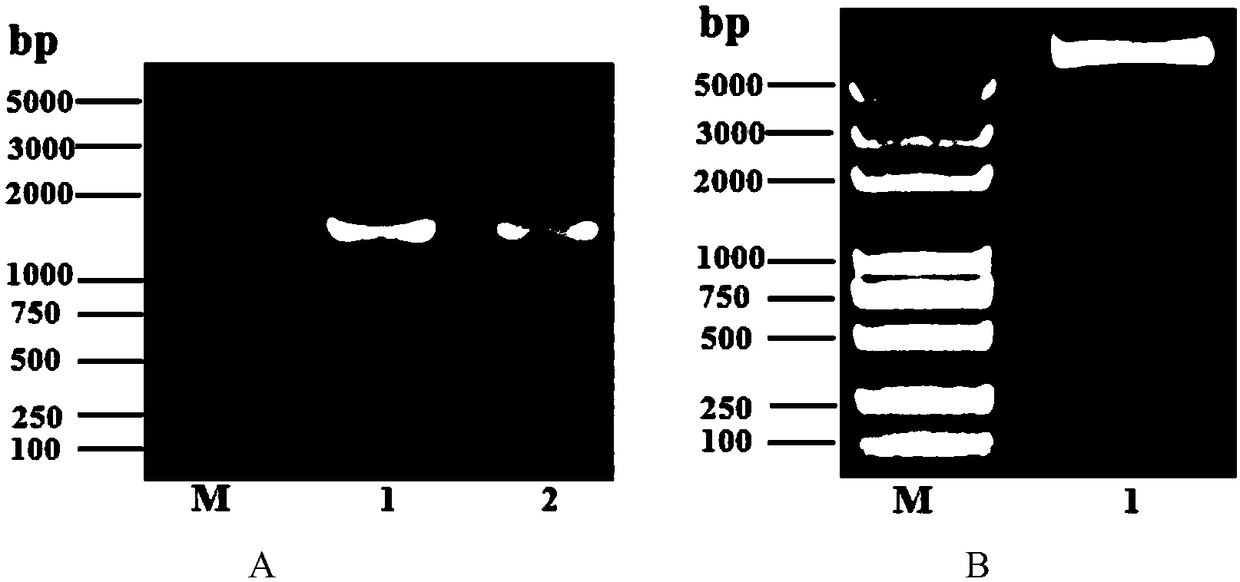
[0031] The SacB gene was amplified using the plasmid pEX18GM (Gene.1998; 212(1):77–86) as a template, and the primers were SacBP1: 5'-catgccatggcaatgaacatcaaaaagtttgc-3' (SEQ ID NO.1), SacBP2: 5'-ccgctcgagttatttgttaactgttaattgtcc- 3' (SEQ ID NO.2), the reaction program is: pre-denaturation at 98°C for 30s; denaturation at 98°C for 10s, annealing at 55°C for 30s, extension at 72°C for 1min and 30s, cycled 30 times; final extension at 72°C for 7min, and storage at 16°C. The amplified products were subjected to agarose gel electrophoresis, and the results were as follows: figure 1 shown.
Embodiment 2
[0032] Embodiment 2, test SacB gene as the reverse screening marker experiment of scarless deletion
[0033] The amplified SacB fragment was cloned into the NcoI and XhoI restriction sites of the shuttle plasmid pLMF02 (Sci Rep.2016, 6:37159), and then transformed into R. anatipestife ATCC by conjugative transfer to obtain strain R. anatipestife ATCC pLMF02::SacB; at the same time, the plasmid pLMF02 was transferred into R.anatipestife ATCC to obtain R.anatipestife ATCC pLMF02, and the recombinant plasmid identification results were as follows figure 2 shown. The strains R.anatipestifeATCC pLMF02 and R.anatipestife ATCC pLMF02::SacB were cultured on the blood plate containing sucrose, and R.anatipestife ATCC pLMF02::SacB showed sensitivity to sucrose, while the control R.anatipestifeATCC pLMF02 was not sensitive to sucrose sensitive. This result indicated that the SacB gene can be used for subsequent studies on scarless deletions.
Embodiment 3
[0034] Embodiment 3, sucrose sensitivity test
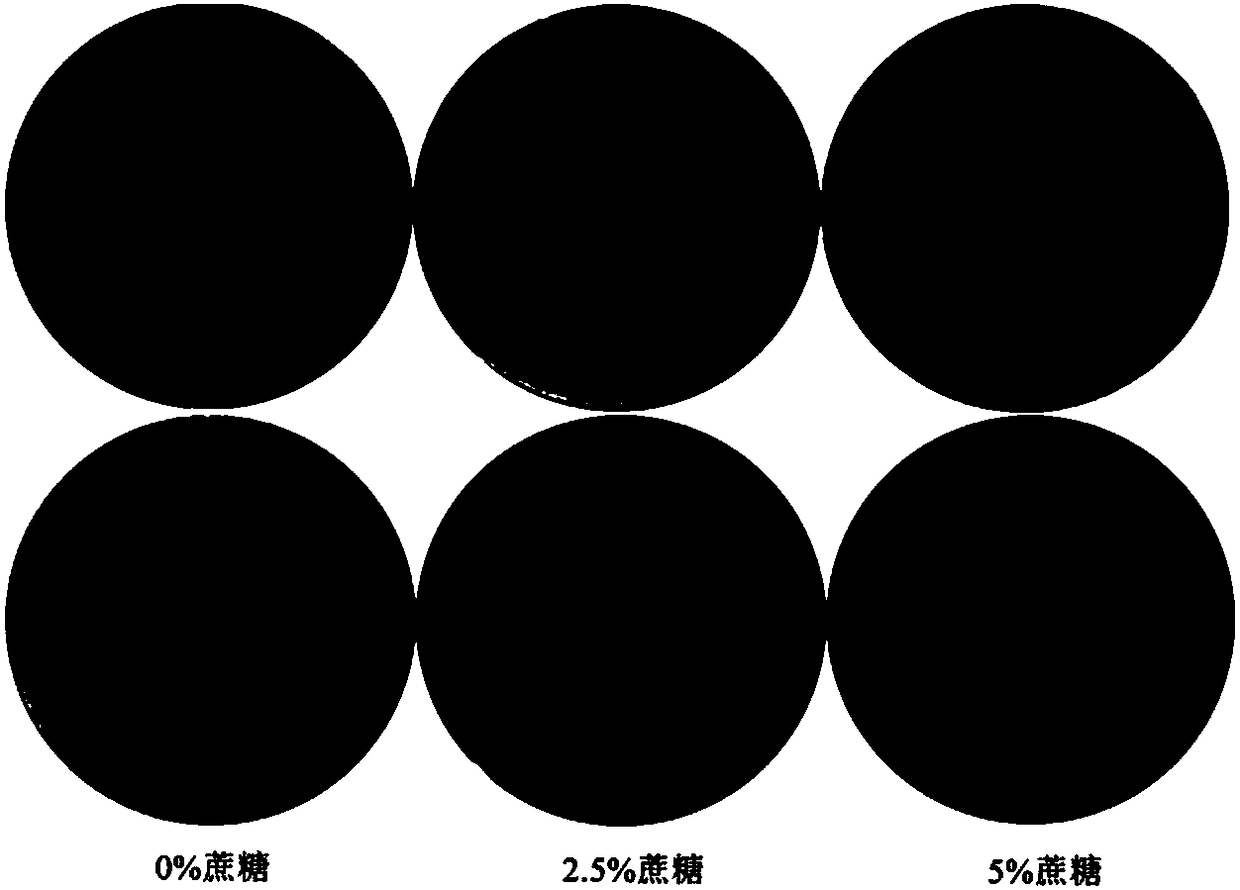
[0035] will contain 10 7 CFU of R.anatipestifer ATCC pLMF02::SacB and R.anatipestiferATCC pLMF02 bacterial solution were applied to the blood plate containing different concentrations of sucrose (0%, 2.5%, 5%), cultivated in a 37°C incubator for 18h, and the growth conditions were as follows: image 3 shown. The results showed that the strain carrying the SacB gene did not grow when the sucrose concentration was 5%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com