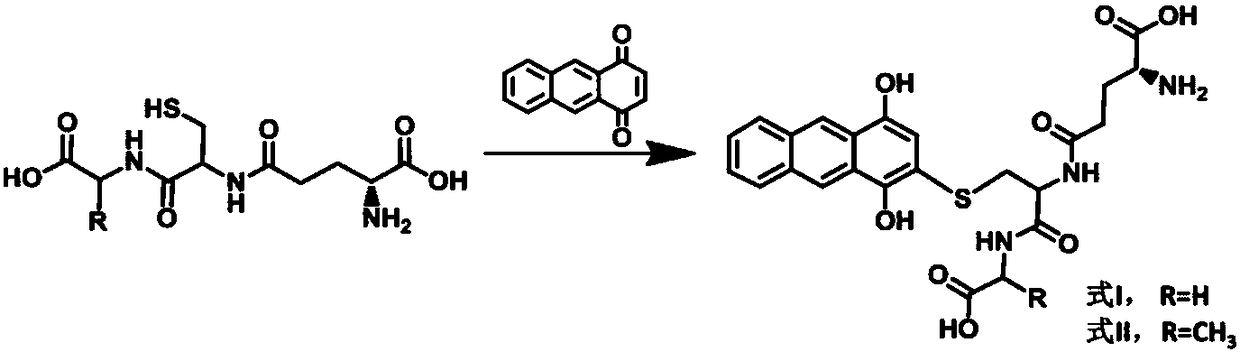
MALDI-TOF mass spectrometric probe for gamma-glutamyl transpeptidase detection and use method thereof
A technology of glutamyl transpeptidase and mass spectrometry, applied in the field of analysis and testing, can solve problems such as synthesis difficulties, autofluorescence interference of biological samples, and photobleaching of molecular probes, and achieve stability under mass spectrometry conditions, easy promotion and application, The effect of good sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Preparation of molecular probes and internal standard molecules of the present invention.
[0045] Under normal temperature conditions, 4mmol glutathione (ECG) and 4mmol tripeptide (ECA) were dissolved in 5mL 50% acetonitrile / water (v / v), respectively, and then 4mmol anthraquinone was added, stirred for 3h, and evaporated under reduced pressure After removing the solvent, use column chromatography to separate and purify to obtain the final product of tan solid, which is formula I and formula II, and its structure confirmation results are as follows:
[0046] Formula I: 1 H NMR(400MHz,298K,DMSO-d6):δ:8.68(m,1H),8.29(m,1H),7.79(m,1H),7.02(s,1H),4.68(m,1H),3.80 (d, 2H, J=6.0Hz), 3.36(d, 2H, J=6.0Hz), 3.15(m, 1H), 2.38(m, 2H), 1.97(t, 2H).ESI-MS: m / z 516.443;
[0047] Formula II: 1 H NMR(400MHz,298K,DMSO-d6):δ:8.68(m,1H),8.29(m,1H),7.80(m,1H),7.02(s,1H),4.07(m,1H),4.24 (m,2H),3.32(d,2H,J=6.0Hz),3.14(m,1H),2.36(m,2H),1.98(t,2H),1.31(d,3H,J=6.0Hz) .ESI-MS: m...
Embodiment 2
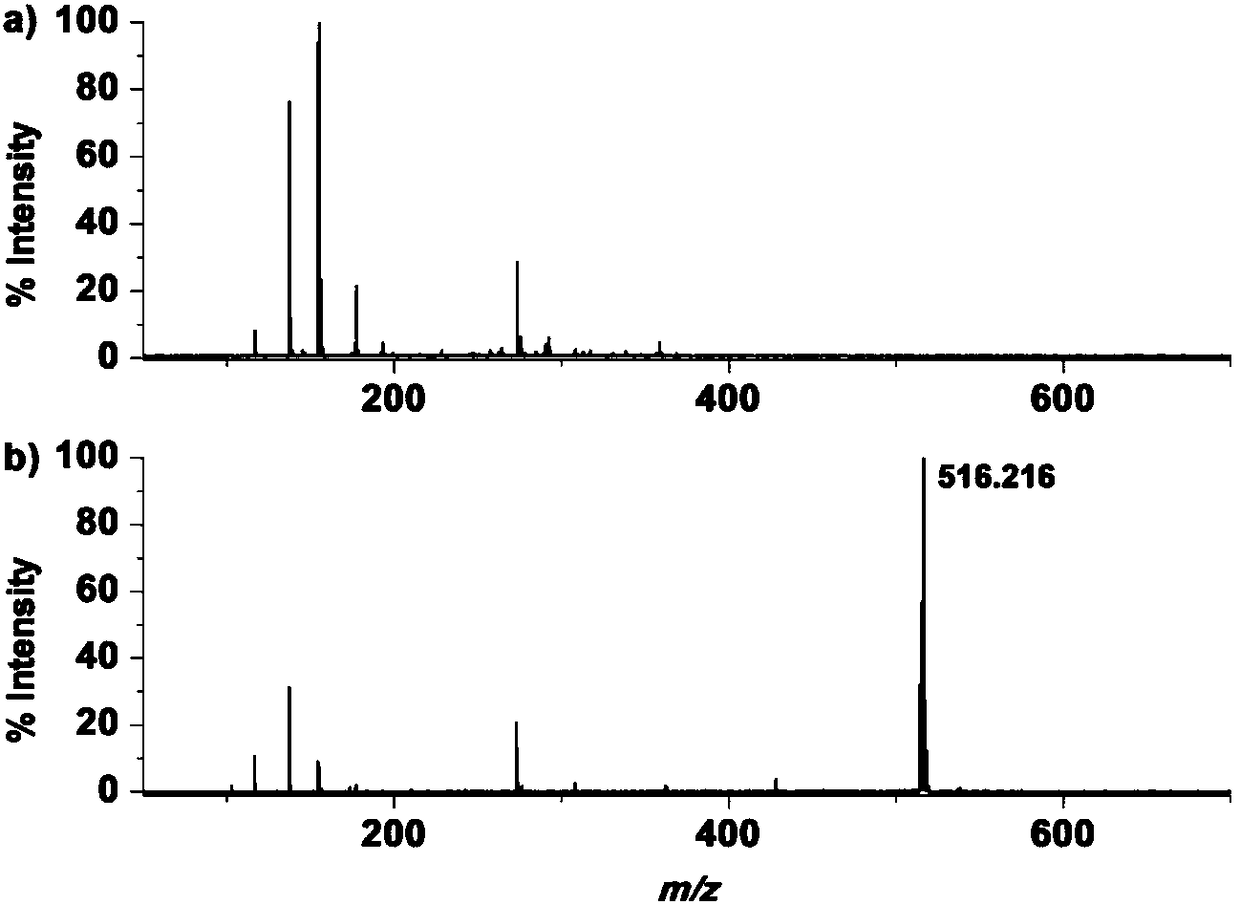
[0048] Example 2: MALDI mass spectrometry analysis of glutathione and the molecular probe of formula I.
[0049] Prepare aqueous solutions of different concentrations of glutathione and molecular probes of formula I, take 1 μL of sample solution and an equal volume of CHCA matrix (5 mg / mL, dissolved in 50% acetonitrile / trifluoroacetic acid aqueous solution (V / V)) or DHB The matrix (20 mg / mL, dissolved in 50% acetonitrile / trifluoroacetic acid aqueous solution (V / V)) was directly mixed on the ground steel target plate, dried at room temperature and analyzed by MALDI mass spectrometry.
[0050] Such as figure 2 Shown are comparison spectra of MALDI assays for 50 μM glutathione and 50 μM Molecular Probe. The peak of glutathione cannot be detected at this concentration, and there are a large number of self-interfering peaks in the traditional matrix, which makes it difficult to identify the peaks and cannot achieve the purpose of accurate characterization. Through the analysis o...
Embodiment 3
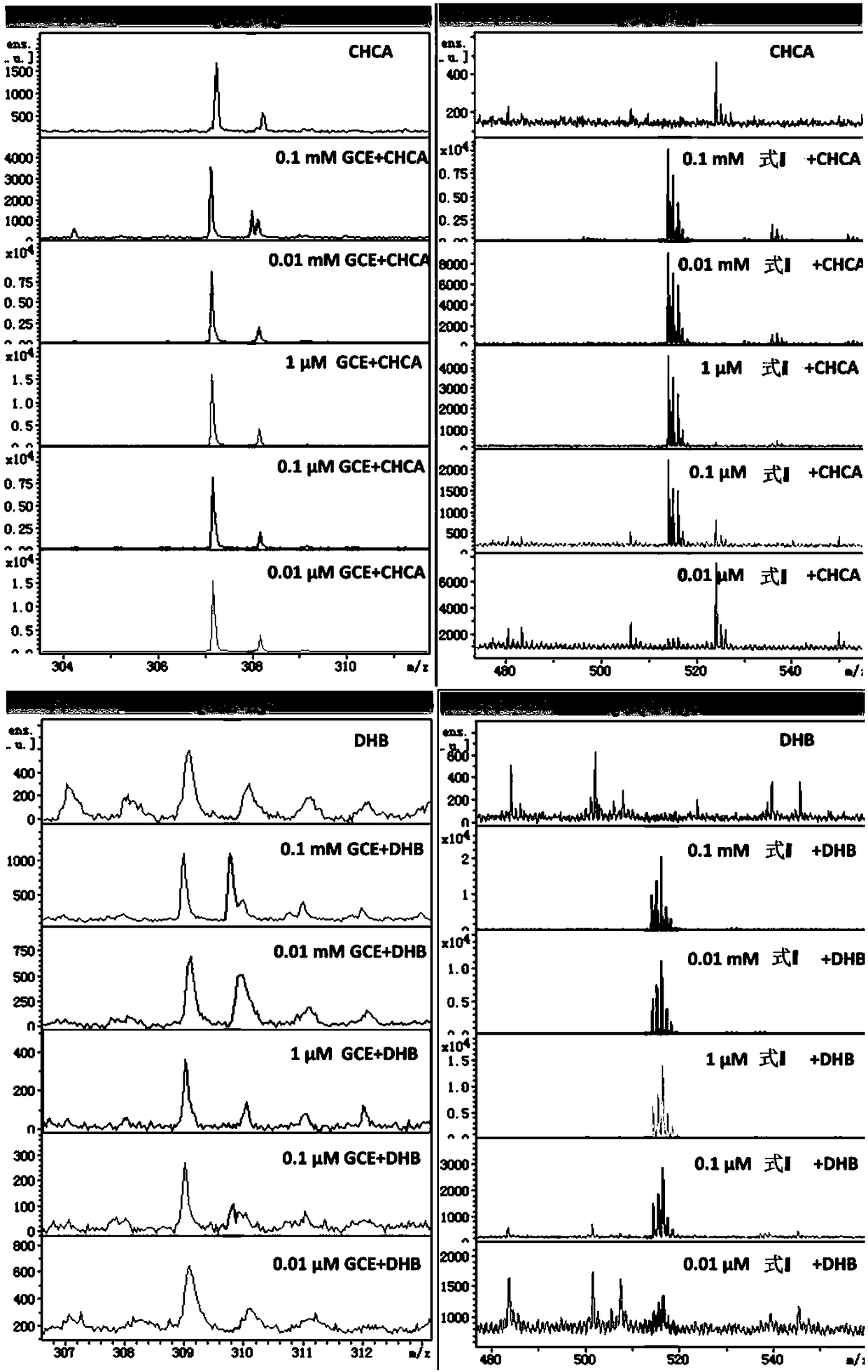
[0051] Example 3: Quantitative analysis of different concentrations of GGTase and drawing a standard curve.
[0052] Configure 500 μM MALDI formula I molecular probe and internal standard aqueous solution as stock solution;
[0053] Configure 180 μL of GGT enzyme standard PBS buffer solution (155.2mM NaCl, 2.97mM, Na 2 HPO 4 ,1.05mM KH 2 PO 4 ; pH=7.4), 20 μL of the stock solution of molecular probes was added, and the digestion reaction was carried out in a water bath at 37° C. for 1.5 h. Add 0.5 μL of formic acid solution and 8 μL of internal standard solution, mix well, take 1 μL of sample solution and 1 μL of DHB matrix solution and mix directly on the ground steel target plate, dry at room temperature and perform MALDI mass spectrometry analysis. Value the abscissa with the different concentrations of GGT enzyme, account for the ratio (I II / I I +I II ) is the ordinate, and the standard curve is drawn.
[0054] from Figure 4 It can be seen that the molecular pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com