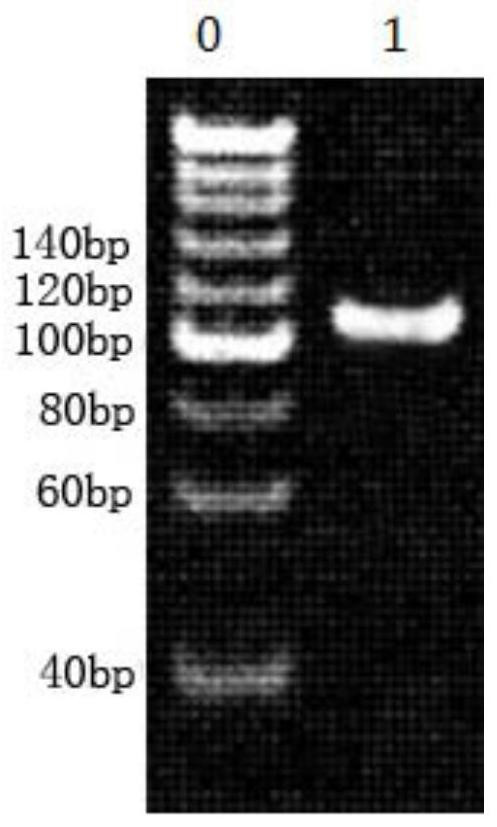
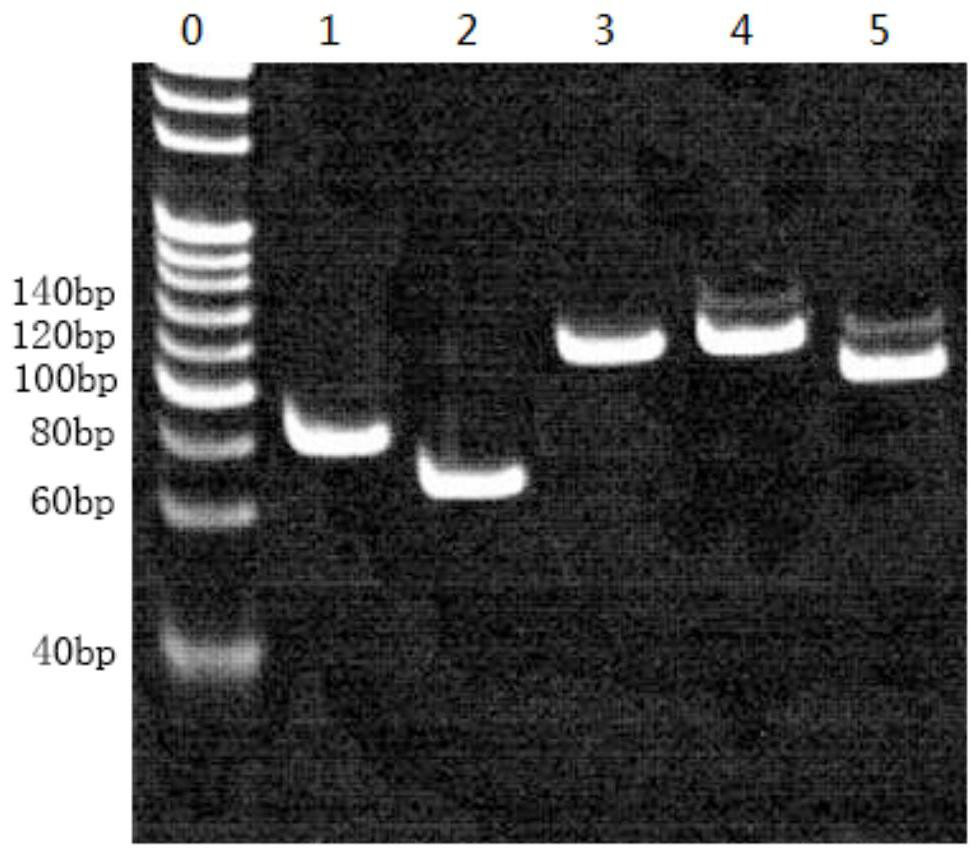
A method for building a library and a method for SNP typing
A typing method and library technology, applied in the field of molecular biology, can solve problems such as the influence of SNP typing accuracy and the mutual interference of different sequencing primers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
no. 1 example
[0023] The present invention proposes a first embodiment, a method for building a database, including the following steps:
[0024] A. Use the specific amplification primer set to perform PCR amplification on the sample to be sequenced containing the SNP site to be detected to obtain the amplified product; at least one of the amplification primers in the specific amplification primer set contains IIS type A restriction endonuclease recognition sequence, the amplified product contains an IIS type restriction endonuclease cleavage site, the distance between the IIS type restriction endonuclease cleavage site and the SNP site to be detected is 0 to 5 bases;
[0025] B. Digest the amplified product with an IIS type restriction endonuclease to obtain a first nucleic acid fragment containing the SNP site to be detected, and the first nucleic acid fragment is digested to form a first end;
[0026] C. Under the action of ligase, the first nucleic acid fragment is ligated with a seque...
no. 6 example
[0075] The present invention also proposes a sixth embodiment, a SNP typing method, including the step of sequencing the library molecules prepared by the library construction method in any of the above embodiments.
[0076] Because the library molecule of the present invention contains sequencing adapters, the sequencing primers of multiple different SNP sites to be tested in the same system are unified, and mutual interference between sequencing primers is avoided.
[0077] Preferably, the method further includes the step of addressably immobilizing library molecules on a solid support.
[0078] The addressable fix refers to a fix that can determine location information. That is, the library molecules immobilized at each specific position on the solid phase support can be clearly distinguished from the library molecules immobilized at other positions.
[0079] Furthermore, the library molecules containing the sequencing adapters can be addressably immobilized on the solid p...
no. 7 example
[0089] The present invention also proposes a seventh embodiment, a method for detecting the rs1801133 site of the MTHFR gene. This embodiment further includes the following steps on the basis of the second embodiment:
[0090] Add 20 μL of 0.1M NaOH solution to the library molecules adsorbed on the magnetic beads obtained in step C to denature the template into a single strand, separate and remove the supernatant, and use 20 μL of 1×TE (containing 0.01% triton) was washed twice, 20 μL 1×TE was washed once, and finally resuspended in 10 μL 1×TE as a sequencing template; the high-throughput gene sequencer Pstar IIA of Shenzhen Huayinkang Gene Technology Co., Ltd. was used for ligation sequencing For sequencing, the 5' end of the sequencing primer (SEQ IDNO:12) is modified with phosphoric acid, and fixed on the sequencing primer binding sequence of the sequencing adapter, and the degenerate nonamer NNNNXNNNN with a fluorescent group complementary to the detection site is used as t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com