Use of a genetic signature diagnostically to evaluate treatment strategies for prostate cancer
A prostate cancer, gene technology, applied in disease diagnosis, microbial determination/examination, instruments, etc., can solve problems such as different responses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0239] Example 1: Development and evaluation of a genomic classifier to predict hormone therapy failure.
[0240] A genomic classifier for predicting hormone therapy failure in prostate cancer subjects was developed as follows. Neuroendocrine prostate cancer (NEPC) can be less sensitive or even resistant to androgen deprivation therapy (ADT), one of the main treatment options for locally advanced, recurrent and metastatic prostate cancer. We collected a set of 1,557 genes to construct a genomic classifier that predicts response to ADT.
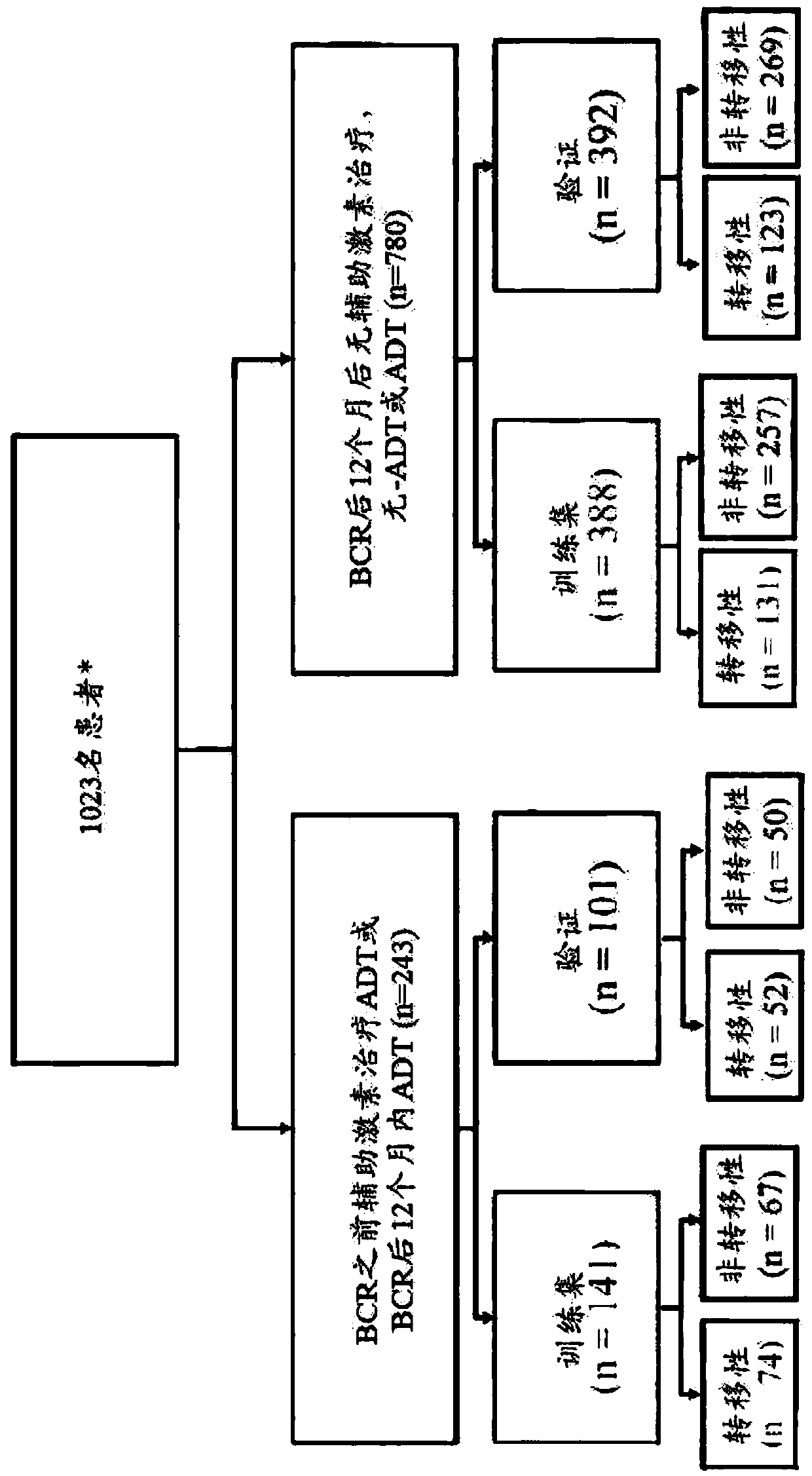
[0241] A total of 529 subjects' prostate cancer expression profiles from formalin-fixed paraffin-embedded (FFPE) tissues were analyzed from the Decipher GRID database and used as a signature for developing androgen withdrawal therapy (ADT) resistance ( ARS) training set. The training data was collected from a nested case-control design cohort in the Mayo Clinic tumor registry, which was previously used to develop the Decipher prostate cancer...
example 2
[0254] Example 2: Systems biology of the ARS gene
[0255] Functional characterization of genes performed in the ARS model revealed that the genes are involved in multiple biological pathways and biological processes involved in cancer progression and neuronal development ( Figure 5 ). Several genes such as NSMCE4A, FOXM1, TFAP, GABRB3 and ATF5 are associated with DNA damage response and apoptosis signaling pathways. In addition, some of the genes have evidence of involvement in androgen signaling. For example, HEY2 has been reported to specifically inhibit AR-dependent transcriptional activity (Villaronga et al. (2008) Curr. Cancer Drug Targets 8(7):566-580). Other ARS genes, including RRM2 and CRISP2, play a role in the cell cycle and cell proliferation. RRM2 plays a role in the proliferation, invasion of prostate cancer, and when it is overexpressed, it is associated with the resistance of other cancers to gemcitabine and platinum-based chemotherapy (Wang et al. (2014) ...
Embodiment 3
[0256] Example 3: Independent Validation of ARS Predicting Hormone Therapy Failure After Adjuvant Hormone Therapy
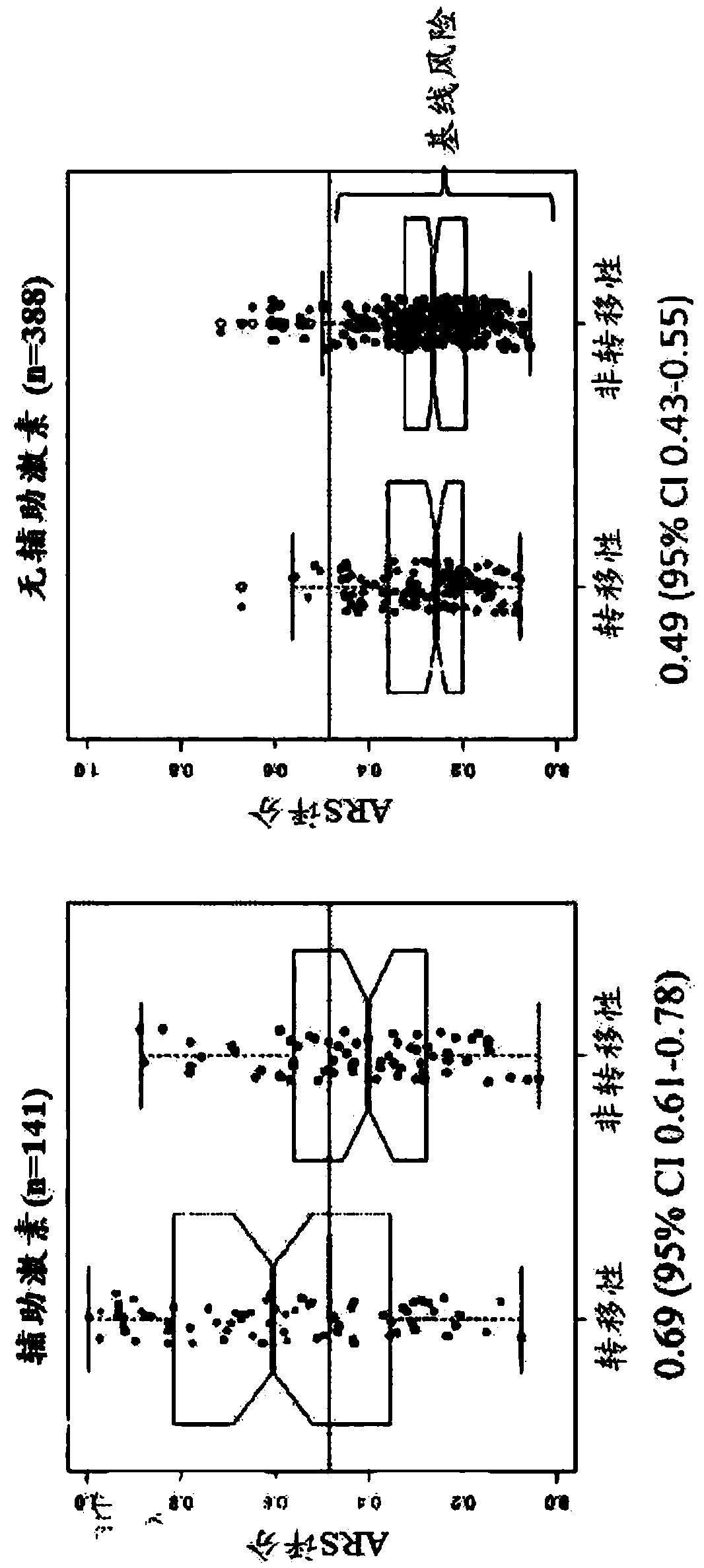
[0257] The ARS model was validated in an independent cohort from Mayo Clinic II (n=233) (Karnes et al. (2013) J. Urol. 190(6):2047-2053; incorporated herein by reference). The cohort was a case cohort design with 101 treated subjects, 51 subjects with metastases, and 132 subjects who did not receive ADT (which had 24 subjects with metastases) (Karnes et al. , ibid.). The same tissue selection and sample processing as in Example 1 was applied. In this independent validation, ARS had a 10-year survival c-index of 0.69 (95% CI 0.59-0.78) in subjects treated with adjuvant ADT compared to untreated subjects The 10-year survival c-index was 0.45 (95% CI 0.29-0.61) ( Image 6 ). Treated subjects with metastases had high scores compared to untreated subjects with baseline scores. These results suggest that treated subjects with higher ARS scores are prone to treatme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com