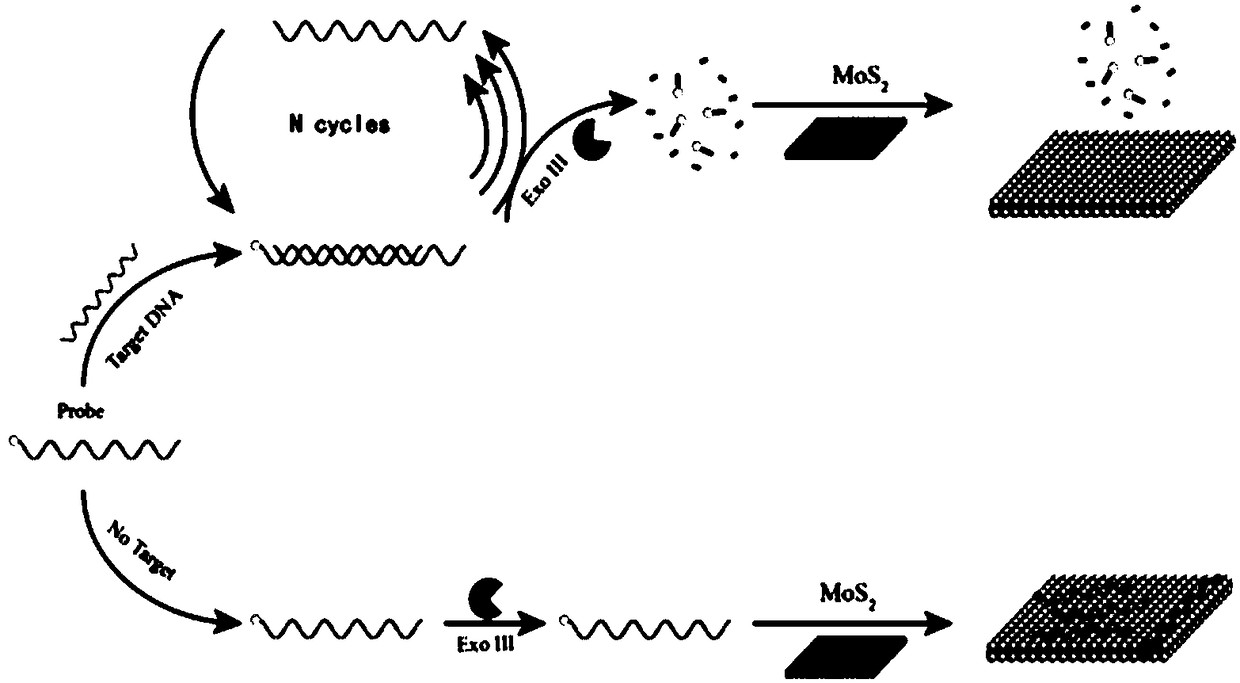
Detection system of nucleic acid and detection method and application thereof
A system and nucleic acid technology, applied in the biological field, can solve problems such as harsh reaction conditions, high design requirements, and poor specificity, and achieve the effects of simple operation, high sensitivity, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] (1) Mix the nucleotide sequence such as the target DNA of SEQ ID NO.2 with the concentration of 5nM and the fluorescent probe of 10nM nucleotide sequence such as SEQID NO.1 and dissolve in 1×NE buffer 1 (New England Biolabs , buffer solution of Exonuclease III), react at a temperature of 25° C. for 10 min, then add 40 U of Exonuclease III to the reacted solution, and heat to inactivate the enzyme after 30 min of enzymatic digestion reaction;
[0059] (2) Compared with step (1), except that no target DNA is added, other conditions are the same as step (1);
[0060] (3) Compared with step (1), except that no fluorescent probe is added, other conditions are the same as step (1);
[0061] (4) Compared with step (1), except that exonuclease III is not added, other conditions are the same as step (1);
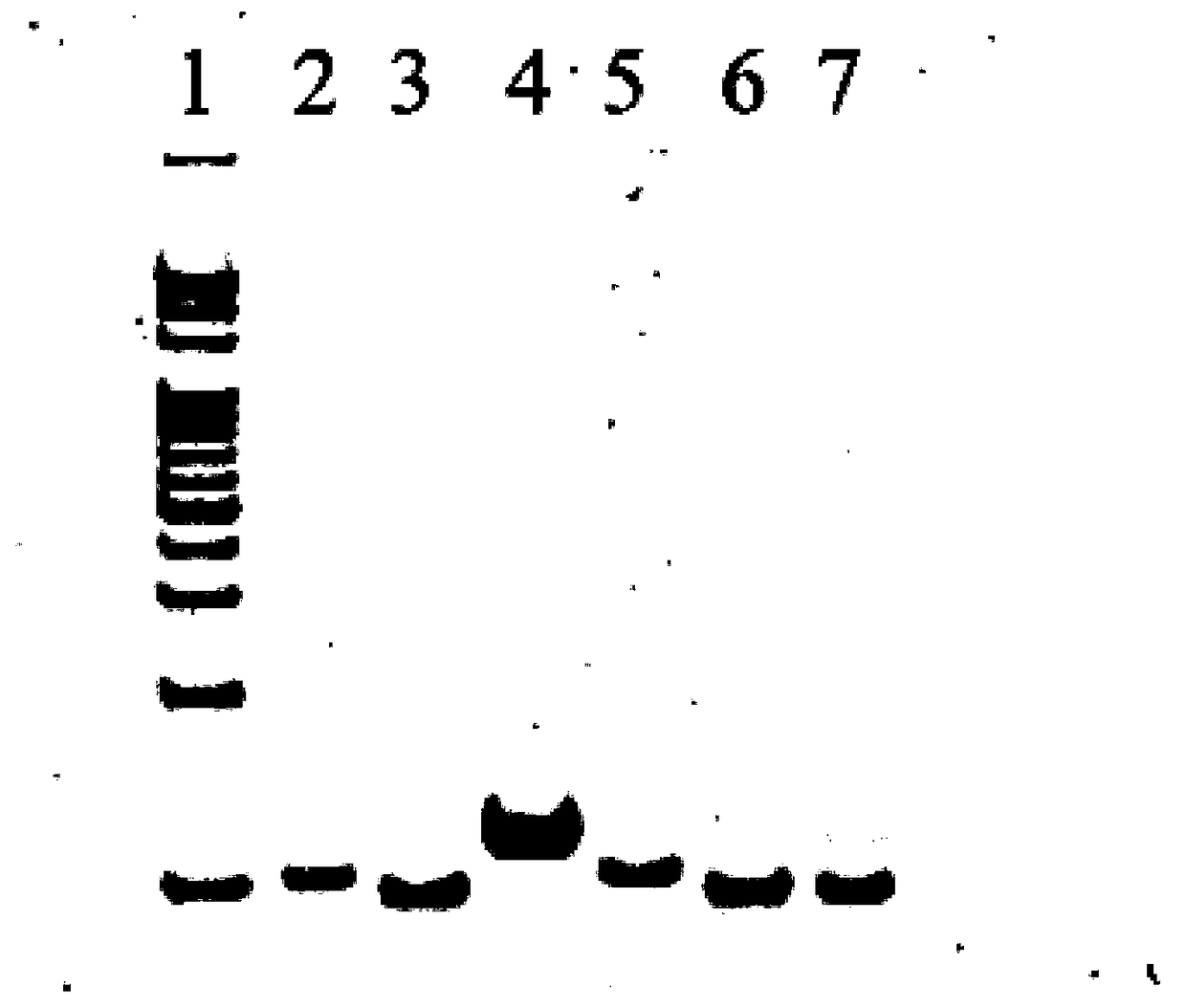
[0062] The post-reaction solutions obtained in steps (1), (2), (3) and (4), as well as separate target DNA and fluorescent probes are loaded separately for polypropylene gel ...
Embodiment 2
[0070] First, optimize the concentration of monolayer molybdenum disulfide nanosheets;
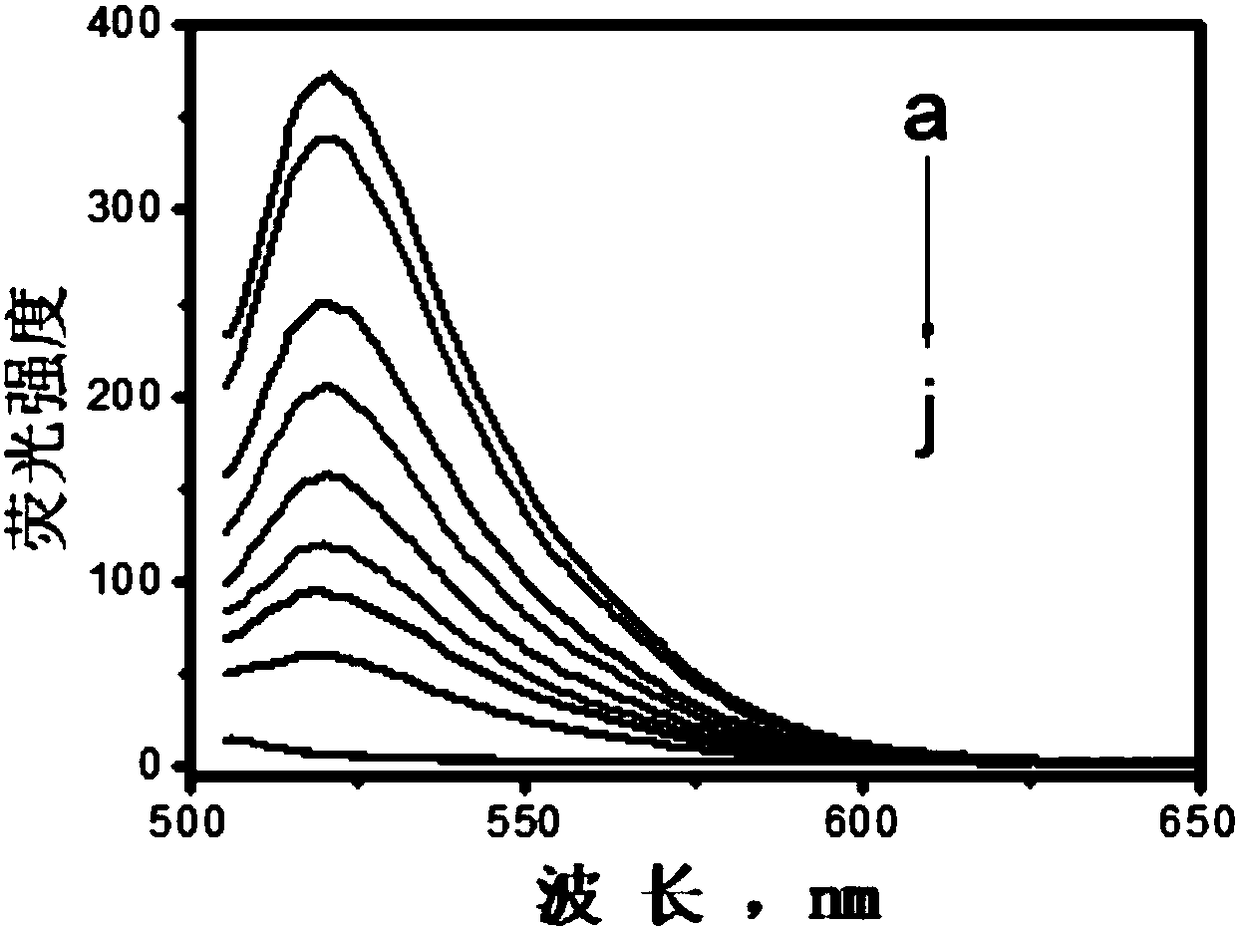
[0071] First, 10 nM of a fluorescent DNA probe with a nucleotide sequence such as SEQ ID NO.1 was dissolved in a Tris-HCl buffer solution, and then a fluorescence spectrometer was used to record its fluorescence spectrum at 505-650 nm at an excitation wavelength of 490 nm;
[0072] Add aliquots (10 μg / mL) of single-layer molybdenum disulfide nanosheets to the above solution nine times, and record the fluorescence spectrum of the tested sample for each addition. The results are shown in Figure 3(A) and Figure 3(B) ;
[0073] It can be seen from Figure 3(A) and Figure 3(B) that with the continuous increase of the concentration of monolayer molybdenum disulfide nanosheets, the fluorescence of the 10nM probe is gradually quenched, when the concentration of monolayer molybdenum disulfide nanosheets is higher than At 80 μg / mL, the fluorescence signal was almost no longer weakened, and the quenc...
Embodiment 3
[0075] Secondly, optimize the amount of exonuclease III enzyme and reaction time;
[0076] First, set up nine large experimental groups, the reaction system contains 0U, 10U, 20U, 30U, 40U, 50U, 60U, 70U and 80U of enzyme, and each experimental group includes two groups, namely the experimental group and The control group, the experimental group and the control group all contain three reaction tubes. The control group does not contain a nucleotide sequence such as the target NDA of SEQ ID NO.2, while the experimental group contains a nucleotide sequence such as the target NDA of SEQ ID NO.2. Target DNA, other conditions remain unchanged, signal-to-noise ratio data = reaction group ÷ control group;
[0077] Secondly, set up six large experimental groups, and the reaction times of adding enzymes into the system are 0min, 10min, 20min, 30min, 40min and 50min, respectively. Each large experimental group contains two groups, the experimental group and the control group, and the exp...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com