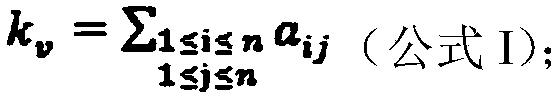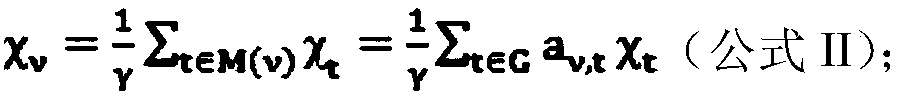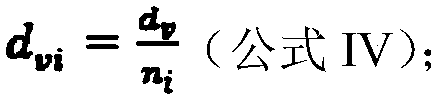Method for identifying esophageal squamous cell carcinoma markers on basis of network index difference analysis
A technology of esophageal squamous cell carcinoma and identification method, applied in the field of bioinformatics, can solve problems such as low accuracy, inability to identify gene expression differences, and easy to be affected by data noise, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Based on the above formula, a method for identifying markers of esophageal squamous cell carcinoma based on network index difference analysis of the present invention will be described below in conjunction with specific embodiments, including the following steps:
[0041]Step 1: download the data set numbered GSE23400 in NCBI, which includes data sets from two different chips (Affymetrix U133A / B chip), the present invention selects the probe data on chip B for data processing, chip B It contains 51 esophageal squamous cell carcinoma sample data and corresponding 51 normal sample data (two interfering probe data are discarded in each sample). The processing of the two sample data is as follows:
[0042] (1) use Expression Console TM Software standardizes the data;
[0043] (2) Convert each data to P-value and robust average signal value by MAS5.0 method;
[0044] (3) Delete the probe data without corresponding genes, and then filter out the genes that are hardly exp...
Embodiment 2
[0098] Example 2 Reliability Verification of Gene Interaction Network
[0099] In order to verify the reliability of the established network relationship, the expression data of some samples were randomly selected, and the Spearman correlation between each gene was still calculated, and a repeatability test was performed. If most of these network relationships can still be found in the subset of samples, it shows that the network relationships we have established are relatively reliable.
[0100] 45 gene sample data were randomly selected from the two sample data of the experimental group, and the intergenic correlation coefficient was calculated according to the gene expression array, and all relationship pairs with a correlation coefficient>=0.8 were screened out as the verification group, and compared with the experimental group. The relationship pairs were compared, and a fixed number of gene relationship pairs were taken for each, and the repetition rate was calculated. R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com