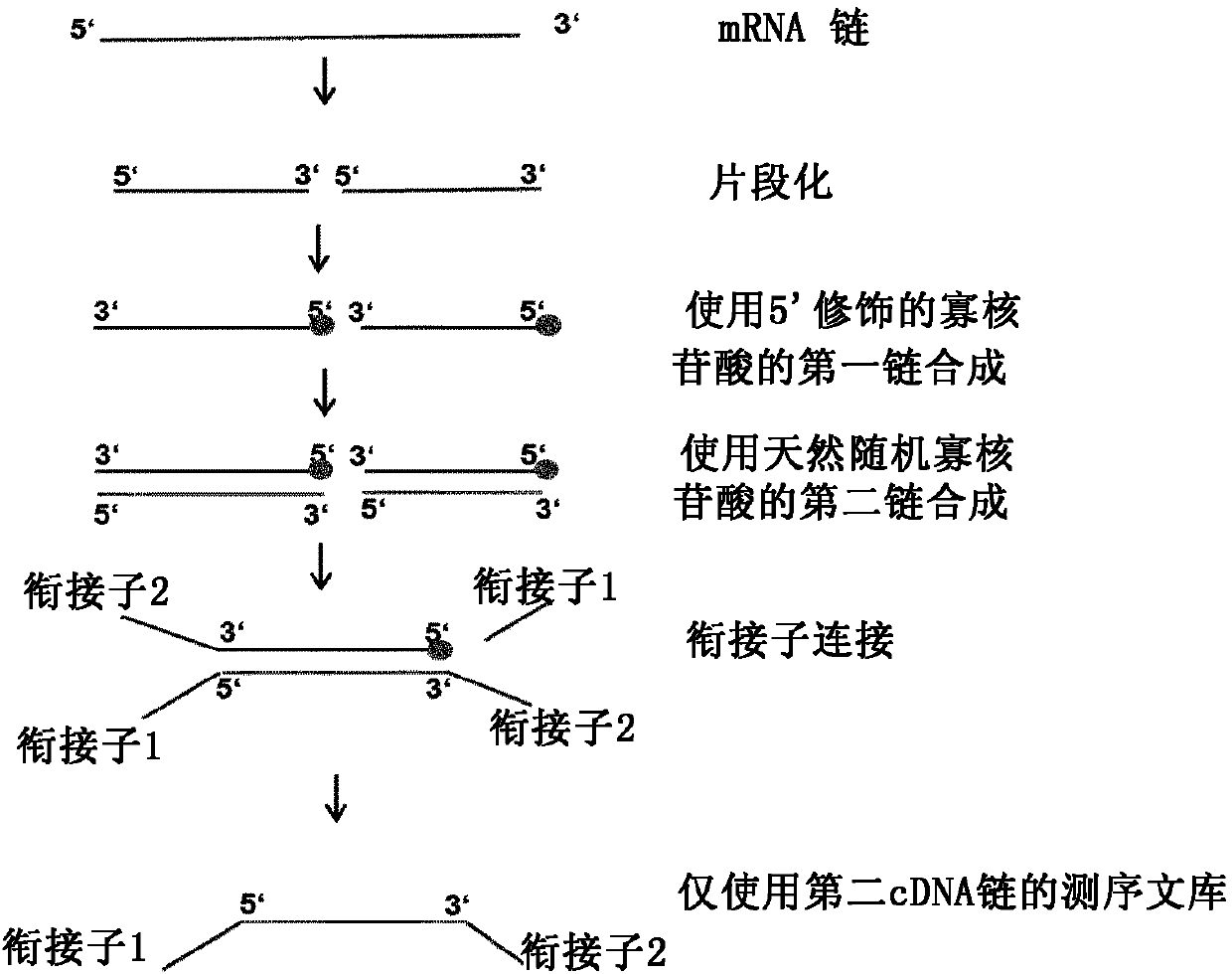
Method for generating a RNA-sequencing library
A technology for sequencing, DNA strands, applied in the field of preparing strand-specific RNA sequencing libraries, which can solve problems such as time-consuming, UNG reaction is not 100% effective, and misinterpretation of RNA sequencing data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0191]RNA from HeLa cells was extracted with RNeasy kit (QIAGEN), and PolyA+mRNA was enriched with GeneRead Pure mRNA Kit (QIAGEN). 86ng of PolyA+ mRNA was then used in each RNA-Seq library preparation reaction according to the following protocol:
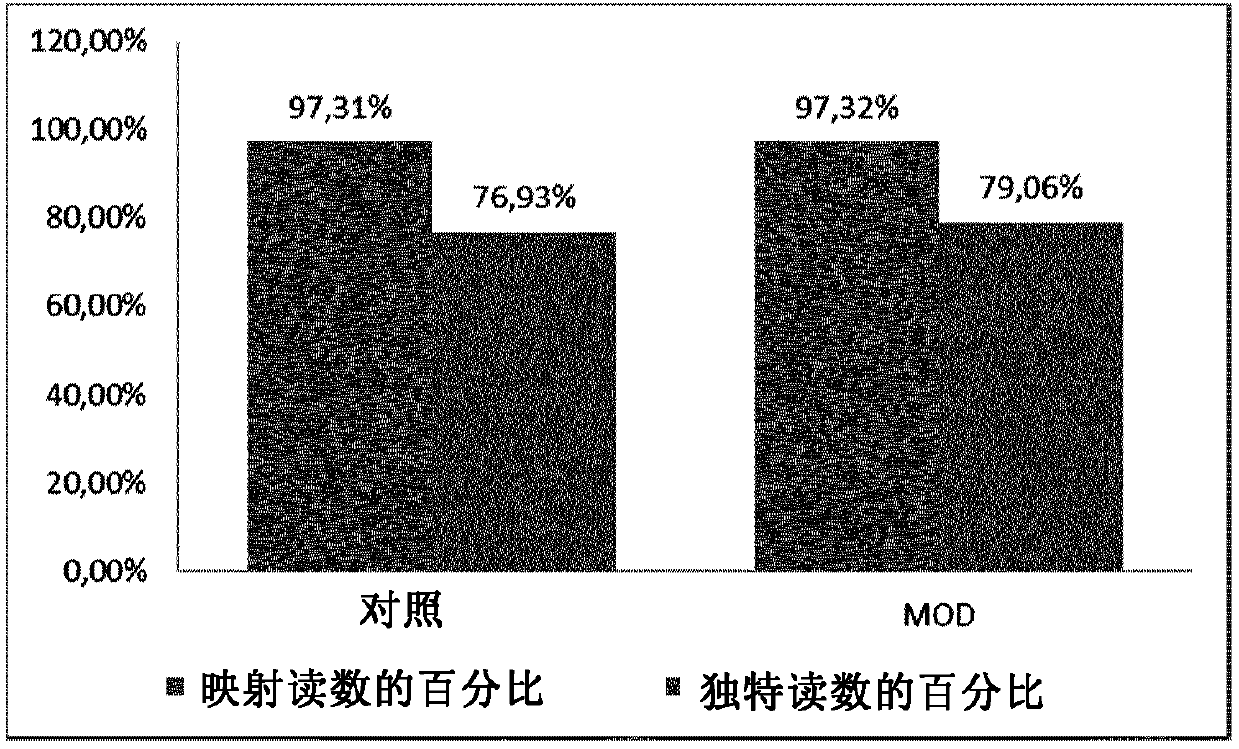
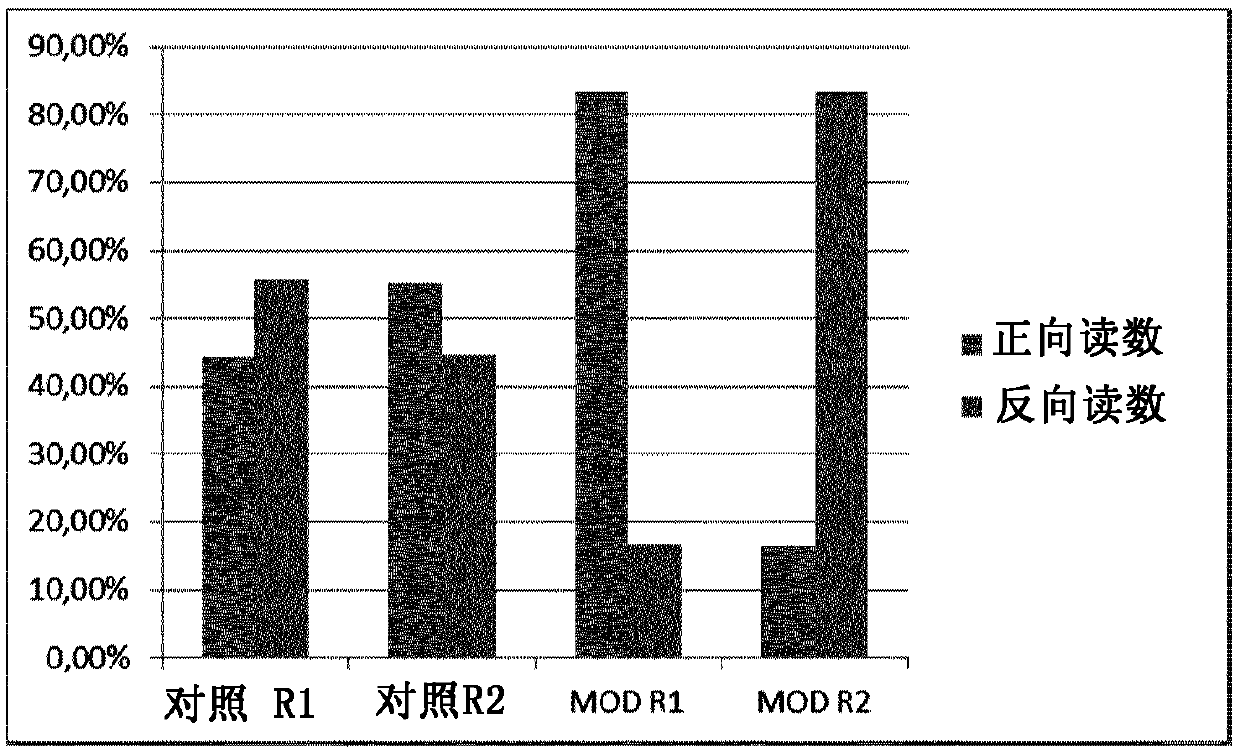
[0192] 32 μl of mRNA (total amount: 86 ng) was mixed with 8 μl of qScript Flex Reaction Mix (5X) (QuantaBiosciences), 2 μl of random 8mer oligonucleotides (200 μM, IDT). The random oligonucleotides were natural oligonucleotides ("Control") or oligonucleotides with a C3 spacer at the 5' ('Mod', / 5SpC3 / NNN NNN NN, IDT)), which would block Ligation of the resulting cDNA to the sequencing adapters.
[0193] The mRNA / random oligonucleotide mixture was heated at 94°C for 15 minutes to fragment the RNA to an average size of approximately 100-200 bp. After heat-mediated fragmentation, the mixture was cooled on ice.
[0194] Subsequently, reverse transcription (RT) components were added: 2 μl RNAse inhibitor (4 U / μl, QIAGEN), 2 μl dNTPs ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com