Methods and systems for determining proportions of distinct cell subsets
A technology of cell subgroups and subgroups, applied in biochemical equipment and methods, biostatistics, molecular entity identification, etc., can solve problems such as difficult to explain results, not very effective, and no statistical significance test
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0208] Example 1: Enhanced Enumeration of Cell Subpopulations by Expression Profiling of Composite Tissues
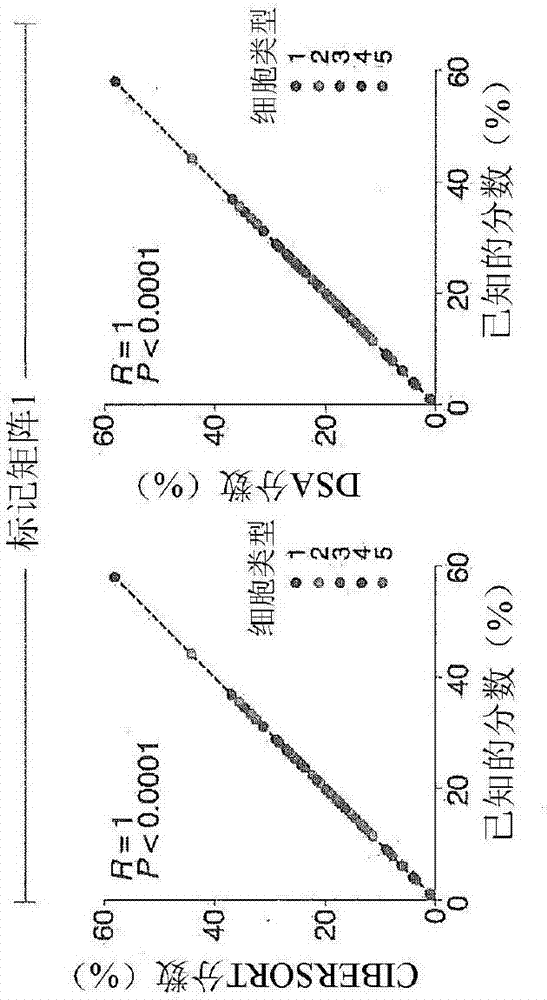
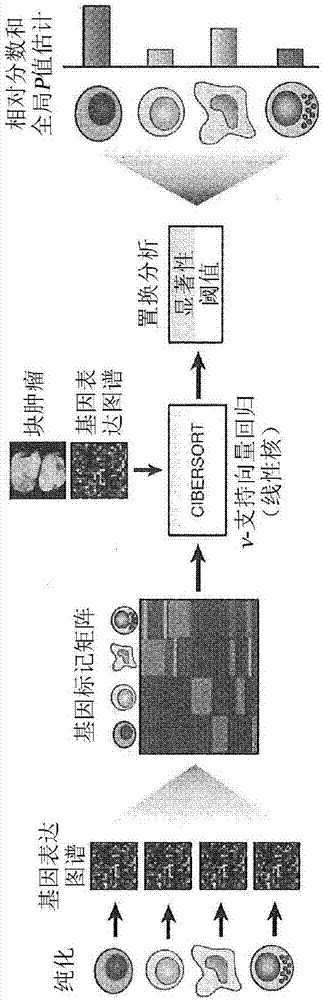
[0209] CIBERSORT uses an input matrix of reference gene expression markers to estimate the relative proportion of each cell type of interest. However, for each gene, no cell type specific expression pattern (method) is required. To deconvolve mixtures, a machine learning method is robust to noise using a novel application of linear support vector regression (SVR) 9 . Unlike many other methods, SVR performs feature selection, where genes from a marker matrix are adaptively selected to deconvolute a given mixture. An empirically defined global P-value for deconvolution is then determined ( Figure 1a ,method).
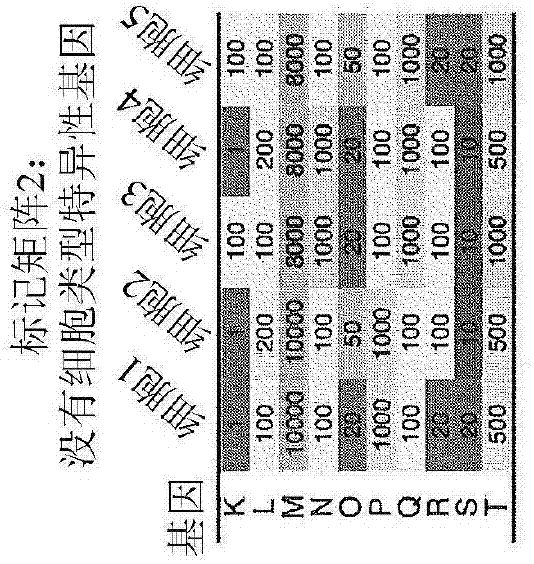
[0210] As a preliminary application, the feasibility of leukocyte deconvolution from bulk tumors, and thus the designed and validated leukocyte marker matrix, was determined. Defined LM22, this marker matrix consists of 547 genes that accurately distinguish...
example 1
[0224] Use the following method for Example 1.
[0225] patient sample
[0226] All patient samples in this study were reviewed and approved by the Stanford Institutional Review Board in accordance with the Declaration of Helsinki. for Figure 5b , with informed consent for research use, at Lucile Packard Children's Hospital, Stanford University, tonsils were collected as part of routine tonsillectomy surgery and then mechanically disintegrated prior to cryopreservation in cell suspension. for Figure 6c "Patient 1" shown in "Patient 1", in subjects without measurable circulatory disease, infused rituximab (375 mg m- 2) Peripheral blood mononuclear cells (PBMC) were isolated from the samples before and immediately after monotherapy. for Figure 6c In patients 2 and 3, PBMCs were isolated from samples collected immediately after 4 and 6 cycles of RCHOP immunochemotherapy for the treatment of DLBCL, respectively. for Figure 6c In patient 4, PBMCs were isolated from a sub...
example 2
[0301] Example 2: Inferred leukocyte frequency and prognostic association in 25 human cancers using CIBERSORT
[0302] Materials and methods
[0303] The following materials and methods were used for Examples 2 and 3.
[0304] Prediction of Clinical Outcomes for Genomic Profiles (PRECOG) Composite and Quality Control. To identify cancer gene expression datasets with corresponding patient outcome data, the NCBI Gene Expression Omnibus (GEO), EBI ArrayExpress, NCI caArray, and Stanford Microarray Databases were queried for the terms survival, prognosis, prognostic, or outcome. A Perl script was implemented to download the processed raw data along with associated annotations. For data in NCBI, array platforms were determined from SOFT format files and corresponding annotation files were retrieved from GEO. From these, ProbeIDs, Genbank accession numbers, HUGO gene symbols and gene descriptions were extracted based on the internal headers of the SOFT annotation files. If thi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com