Method for developing molecular marker interlocked with specific chromosome region of wheat
A technology of molecular markers and chromosomes, applied in the field of agricultural biology, can solve problems such as the difficulty of developing markers for specific chromosome segments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Development of molecular markers tightly linked to wheat powdery mildew resistance gene pmHYM:
[0038] 1. Obtain the sequence information of the molecular markers developed:
[0039] 1) According to the preliminary mapping results of pmHYM (Master's thesis of Zhang Zhiliang), the gene is located between the markers Xgwm611 and Xgwm577, due to Nematollahi et al. (2008, Microsatellite mapping of powdery mildew resistance allele Pm5d from common wheat line IGV1-455, Euphytica, 159, 307-313) Markers Xgwm611 and Xgwm577 were positioned in the chromosome arm of wheat 7BL 0.86-1.00, and the EST sequence positioned on the chromosome arm was used as the original sequence;
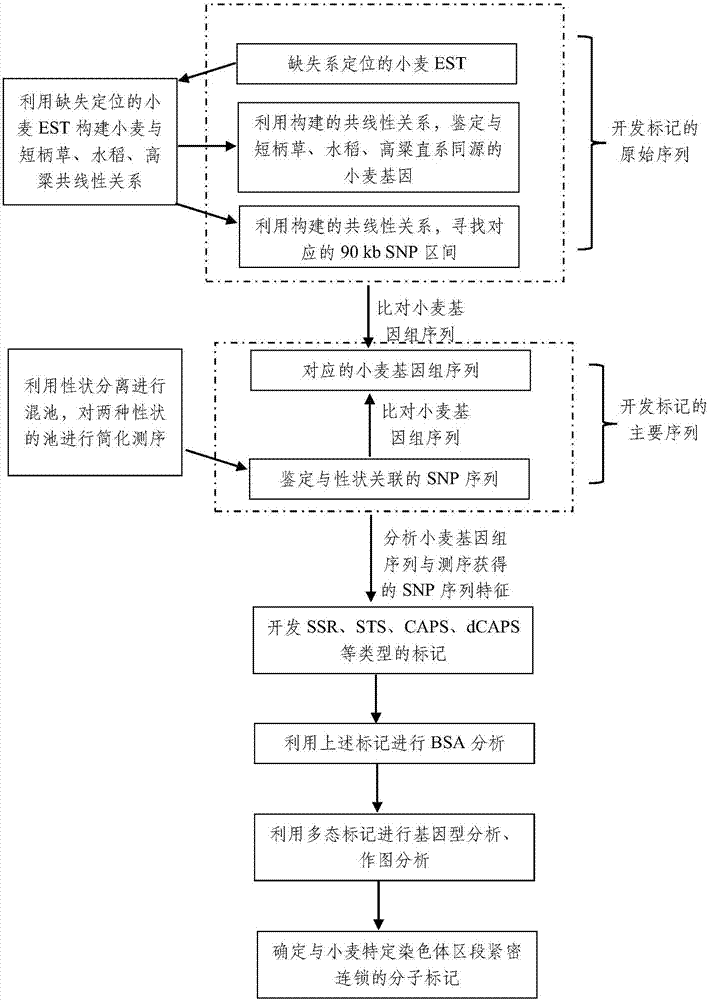
[0040] 2) Use the above-mentioned EST sequences to compare the genomes of Brachypodium, rice, and sorghum, establish the collinearity between this segment and Brachypodium, rice, and sorghum, compare the collinear relationship constructed by the corresponding segment of the 90kb SNP, and determine the Segme...
Embodiment 2
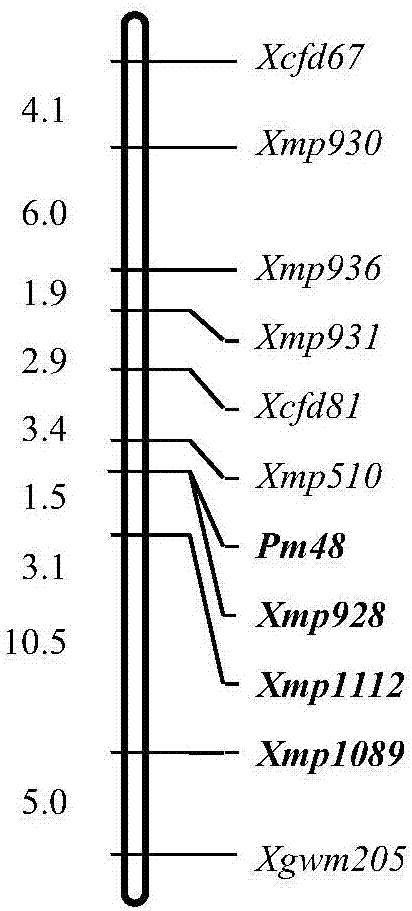
[0048] Development of Molecular Markers Tightly Linked to Wheat Powdery Mildew Resistance Gene Pm48:
[0049] 1. Obtain the sequence information of the molecular markers developed:
[0050] 1) According to the preliminary mapping results of Pm48 (Gao et al., 2012, Genetic analysis and molecular mapping of a new powdery mildew resistant gene Pm46 in common wheat, Theor ApplGenet, 125, 967-973), the gene is located between markers Xcfd81 and Xgwm205, marker Xcfd81 and Xgwm205 may be located in the chromosome arm of wheat 5DS 0.63-1.00, and the EST sequence located on the chromosome arm was used as the original sequence;
[0051] 2) Use the above-mentioned EST sequences to compare the genomes of Brachypodium, rice, and sorghum, establish the collinearity between this segment and Brachypodium, rice, and sorghum, compare the collinear relationship constructed by the corresponding segment of the 90kb SNP, and determine the Segment potential SNP markers, as raw sequences;
[0052] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com