A simple and rapid plant DNA extraction method
An extraction method and plant technology, applied in the field of molecular biology, can solve the problems of high professionalism and carefulness, the influence of material consumption on experimental results, and high requirements for experimenters to operate, so as to reduce human errors, save manpower, and reduce prices. cheap effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
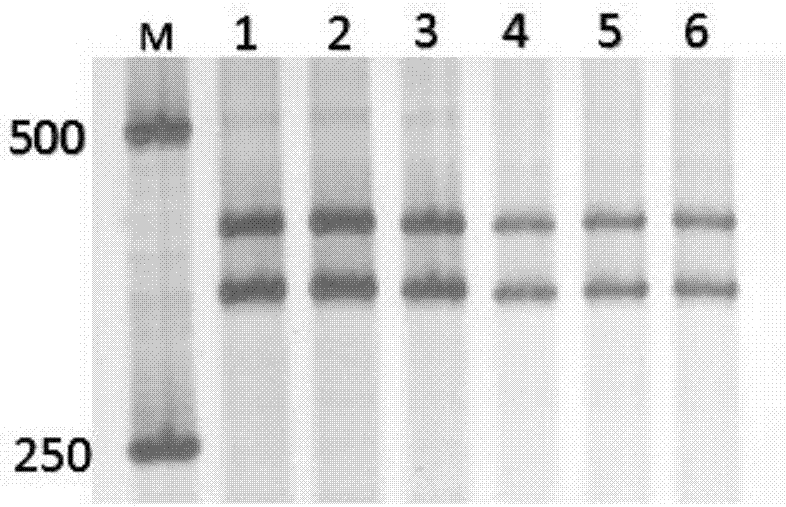
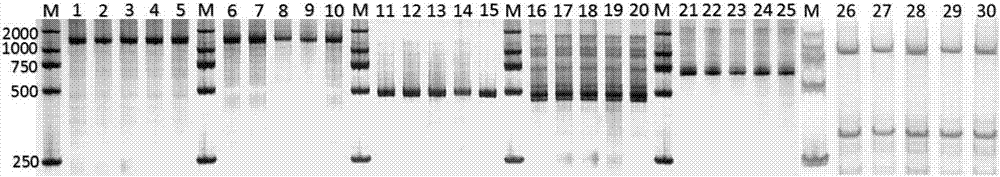
Image
Examples
Embodiment 1
[0031] 1. Preparation of DNA extraction solution
[0032] (1) 0.5M Tris-HCl (200ml): Weigh 60.55g Tris, dissolve it in 100ml distilled water, then adjust the pH to 8.0 with concentrated hydrochloric acid, adjust the volume to 200ml, and sterilize by high temperature steam (121°C, 20 minutes). Store at low temperature (4°C).
[0033] (2) 0.5M EDTA (200ml): Weigh 29.2g of EDTA and dissolve in 100ml of distilled water, then adjust the pH to 8.0 with 10M NaOH, adjust the volume to 200ml, and sterilize by high-temperature steam (121°C, 20 minutes). Store at low temperature (4°C).
[0034](3) Extraction solution (100ml): 10ml 0.5M Tris (pH 8.0), 5ml 0.5M EDTA (pH 8.0), dilute to 100ml, high temperature steam sterilization (121°C, 20 minutes). Store at low temperature (4°C).
[0035] 2. Extraction of DNA
[0036] (1) Take about 1-2cm of fresh wheat leaves 2 In a sterilized 1.5ml centrifuge tube, grind it into a homogenous slurry with a grinding rod. Add 200 μl extract solution ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com