Detection method for common gene mutations in 17α-hydroxylase deficiency applicable to the Chinese population
A crowd, cyp17a1-y329fs technology, applied in biochemical equipment and methods, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0023] Concrete implementation method of the present invention comprises the following steps:
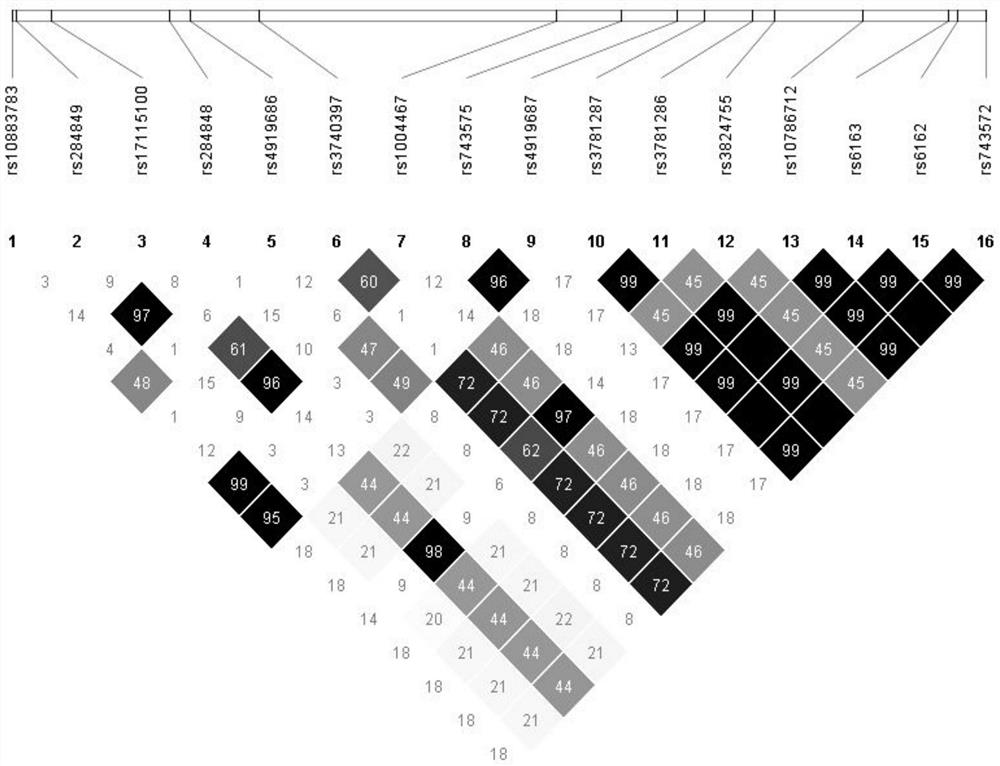
[0024] Step (1): Select 16 SNP sites on the CYP17A1 gene (evenly distributed on the CYP17A1 gene). The selected SNP sites are shown in Table 1.
[0025] Table 1. Selected SNP sites
[0026]
[0027] Step (2): Design 6 pairs of specific primers that can amplify all 16 SNPs. The primers are shown in Table 2. Primer 1F / 1R amplifies SNP1, 2, and 3, primer 2F / 2R amplifies SNP4, and primer 3F / 3R amplified SNP5, 6, 7, 8, primer 4F / 4R amplified SNP9, 10, primer 5F / 5R amplified SNP11, 12, 13, primer 6F / 6R amplified SNP14, 15, 16.
[0028] Table 2. PCR amplification fragments and primers
[0029]
[0030]
[0031] Step (3): Perform gene sequencing on the 16 SNP sites in patients and carriers (Y329fs) to determine the genotype of each site.
[0032] Step (4): Haplotype analysis was performed using PHASE 2.1 software.
[0033] Step (5): Obtain the haplotype haplotype 1-5 containi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com