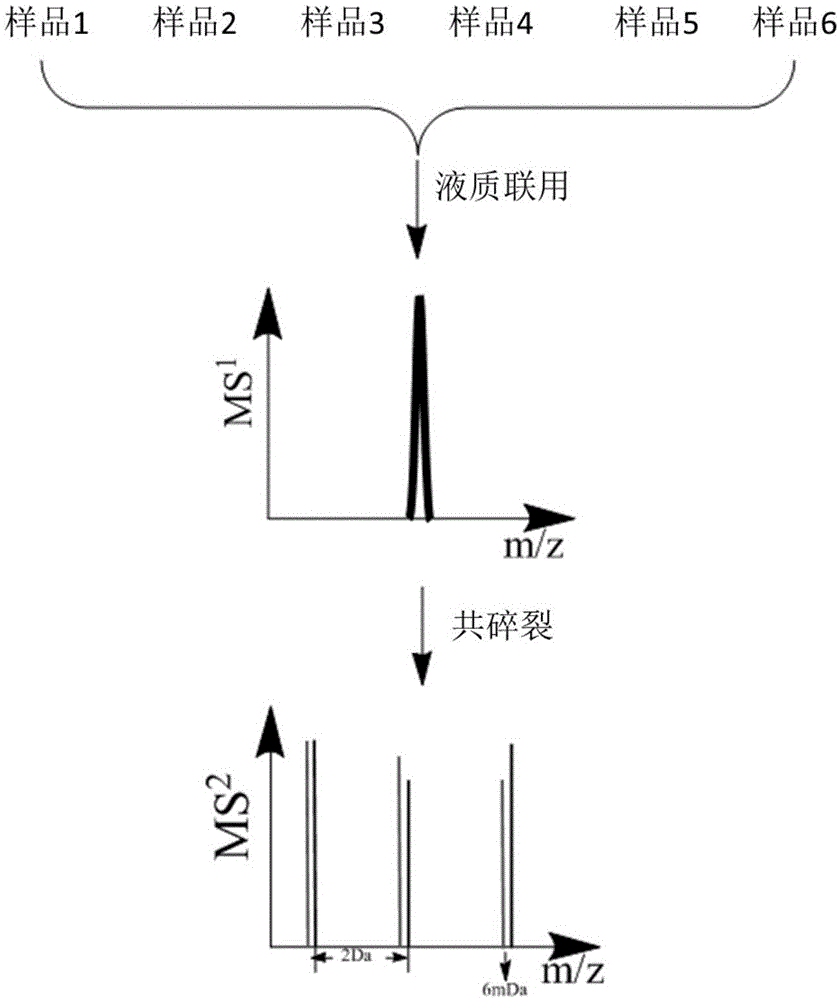
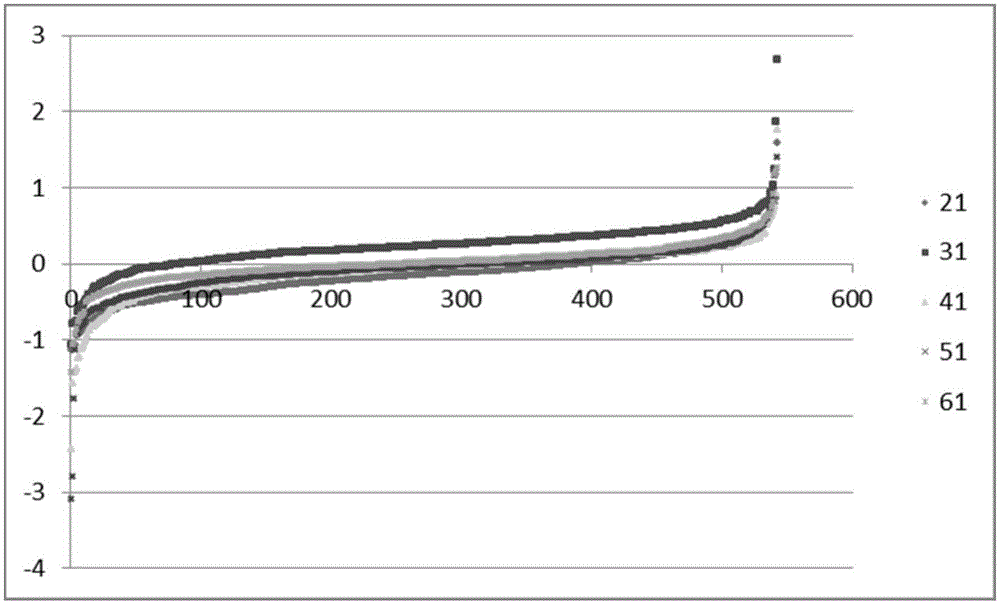
Multiple protein quantification method based on equiponderous dimethylation labeling
A dimethylation and protein technology, applied in the directions of labels, measuring devices, and instruments used in chemical analysis, can solve the problems of small flux and affect quantitative accuracy, and achieve large quantitative flux and improve quantitative accuracy. , Quantitative dynamic range wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
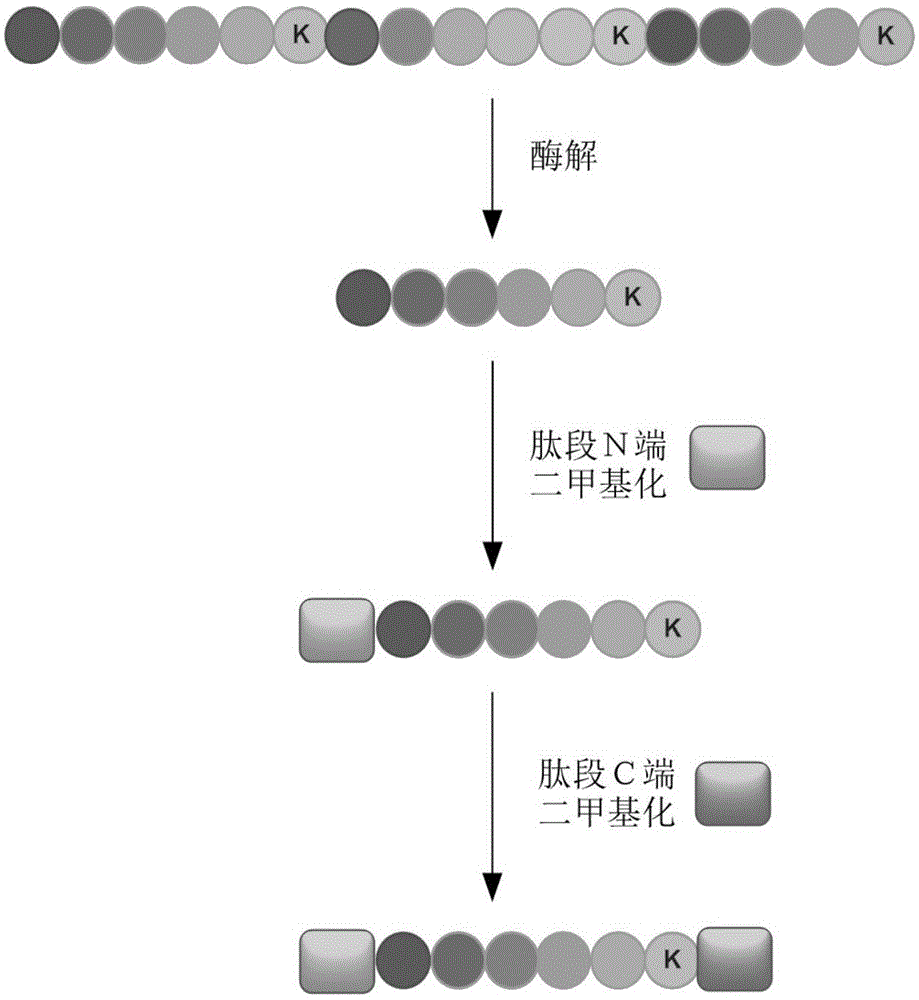
[0030] 1. Protein enzymatic hydrolysis
[0031] Dissolve the protein sample in 8M urea solution, add dithiothreitol to a final concentration of 10mM, denature and reduce at 56°C for 1 hour, add iodoacetamide to a final concentration of 20mM, and react in the dark for 30min, then dilute urea to 1M, Lys-C was added at a ratio of 1:50 (enzyme / protein, w / w) and digested overnight at 37°C. Use a C18 trapping column to desalt and evaporate to dryness, and redissolve in water.
[0032] 2. Peptide dimethylation labeling
[0033] The peptides were marked with six-fold dimethylation, as shown in Table 3. The specific marking method is as follows:
[0034] The first labeling: load the peptide segment after enzymatic hydrolysis onto the C18 trapping column, and first equilibrate the trapping column with an acidic solution with a volume concentration of 1.5% acetic acid aqueous solution. Selectively dimethylate the amino group at the end of the labeled peptide under acidic conditions. T...
Embodiment 2
[0050] The labeling process is carried out in a centrifuge tube. After acidic labeling, solvent exchange is carried out through a trapping column, and basic labeling is performed after elution. Other processes are the same as in Example 1.
Embodiment 3
[0052] The acidic labeling process is carried out in a centrifuge tube, and then the solvent is exchanged through the trapping column, and the basic labeling is performed on the trapping column, and other processes are the same as in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com