A method and kit for accurately quantifying next-generation sequencing libraries with different GC contents using qPCR
A technology of next-generation sequencing library and kit, which is applied in the field of next-generation sequencing library and next-generation sequencing, which can solve problems such as differences, inability to guarantee amplification efficiency, and inability to accurately quantify libraries with different GC contents, so as to avoid quantitative errors , The effect of high-quality sequencing results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
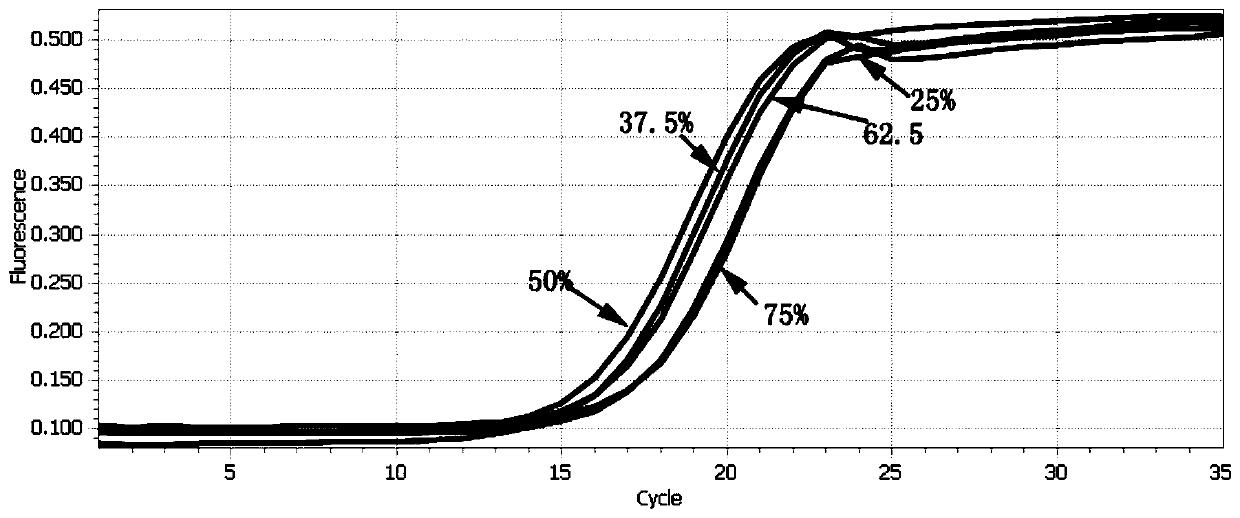
[0055] A kit for accurately quantifying next-generation sequencing libraries with different GC contents by qPCR, including: 5 standards with different GC contents (standards are linear DNA fragments, containing P5 / P7 junction primer sequences, the size of the amplified fragment is 420bp, The GC content is shown in Table 1), library diluent (a mixture of 10mM Tris-HCl with pH=8.0, 1mg / ml BSA and 0.05% Tween20), a new dye-based qPCR master mix TransNGS Green based on antibody-based hot start qPCRSuperMix, primer (Primer Premix), ddH 2 O; wherein, the primer is
[0056] Primer P5:5'-AAT GAT ACG GCG ACC ACC GA-3' (shown in SEQ ID NO:1)
[0057] Primer P7:5'-CAA GCA GAA GAC GGC ATA CGA-3' (shown in SEQ ID NO:2)
[0058] The method for accurately quantifying next-generation sequencing libraries with different GC contents using the above kits includes:
[0059] 1) Selection of standard: According to the GC content of the DNA library to be tested (the genome GC content of common mo...
Embodiment 2
[0068] A kit for accurately quantifying next-generation sequencing libraries with different GC contents by qPCR, including: 7 standards with different GC contents (standards are linear DNA fragments, containing P5 / P7 adapter primer sequences, the size of the amplified fragment is 300bp, The GC content is shown in Table 2), library diluent (a mixture of 10mM Tris-HCl at pH = 8.0 and carrier RNA at a concentration of 0.05mg / ml), a new dye-based qPCR master mix TransNGS Green qPCR SuperMix based on antibody method hot start , primer (Primer Premix), ddH 2 O; wherein, the primer is
[0069] Primer P5:5'-AAT GAT ACG GCG ACC ACC GA-3' (shown in SEQ ID NO:1)
[0070] Primer P7:5'-CAA GCA GAA GAC GGC ATA CGA-3' (shown in SEQ ID NO:2)
[0071] The method for accurately quantifying next-generation sequencing libraries with different GC contents using the above kits includes:
[0072] 1) Selection of standard: According to the GC content of the DNA library to be tested (the genome GC ...
Embodiment 3
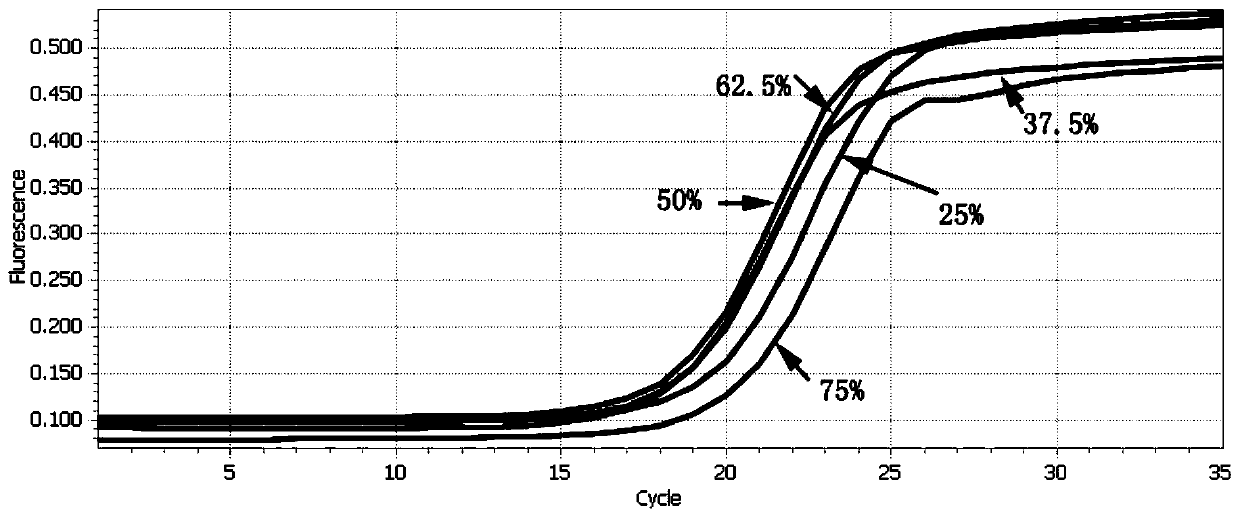
[0081] A kit for accurately quantifying next-generation sequencing libraries with different GC contents by qPCR, including: 5 standards with different GC contents (standards are linear DNA fragments, containing P5 / P7 junction primer sequences, the size of the amplified fragment is 600bp, The GC content is shown in Table 1), library diluent (a mixture of 10mM Tris-HCl with pH = 8.0, EDTA with a concentration of 1mM and 0.05mg / ml tRNA), and a new dye-based qPCR master mix TransNGS based on the hot start of the antibody method Green qPCRSuperMix, Primer (Primer Premix), ddH 2 O; wherein, the primer is
[0082] Primer P5:5'-AAT GAT ACG GCG ACC ACC GA-3' (shown in SEQ ID NO:1)
[0083] Primer P7:5'-CAA GCA GAA GAC GGC ATA CGA-3' (shown in SEQ ID NO:2)
[0084] The method for accurately quantifying next-generation sequencing libraries with different GC contents using the above kits includes:
[0085] 1) Selection of standard: According to the GC content of the DNA library to be t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com