Method for monitoring cancer cell number of cancer patient by adopting circulating DNA
A tumor cell and patient technology, applied in the field of monitoring the number of tumor cells, can solve the problems of the intermediate data detection method and the unsatisfactory effect, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
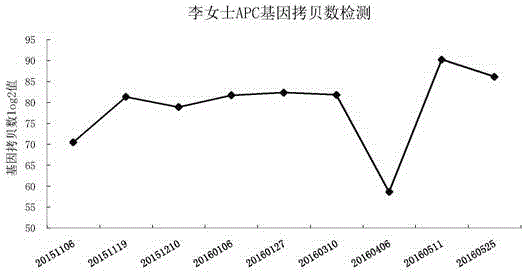
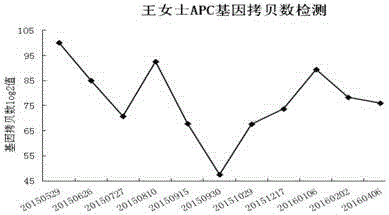
[0041] The following examples will further illustrate the present invention, but do not thereby limit the present invention: 1. Take two patients as examples, and carry out detailed tracking of their therapeutic effects. The first case was a patient with breast cancer and the second case was a patient with non-invasive small cell lung cancer.
[0042] 1. Plasma Separation
[0043] 1) Blood is sucked into a sterile centrifuge tube;
[0044] 2) Immediately start centrifugation, 4°C, 1600g centrifugation for 10min;
[0045] 3) After centrifugation, suck the supernatant into a new centrifuge tube;
[0046] 4) Centrifuge at 16000g for 10min at 4°C;
[0047] 5) Aspirate the supernatant into a 2ml sterile centrifuge tube;
[0048] 6) Store at -20°C.
[0049] 2. ctDNA extraction
[0050] 1) Add 1ml plasma sample to 2ml EP tube, then add 1ml lysis buffer, 30μl proteinase K and 60μl nanoparticle adsorption magnetic beads. Fully invert and mix for 5min.
[0051] 2) Put the mixed ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com