Method for quickly detecting ploidy of new brassica hexaploid germplasm
A Brassica and new germplasm technology, applied in the biological field, can solve the problems of poor effect of very young leaves, unsuitable for large-scale detection, blurred description, etc., to achieve easy judgment, obvious peak bands, and high detection efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] (1) Treat the tender leaf (from the heart leaf to the third tender leaf) sample with a mill to prepare a single-cell suspension:
[0085] 1) At the five-leaf stage of the plant, take about 50 mg of the third tender leaf (the third tender leaf from the heart leaf to the outside) and place it in a centrifuge tube (1.5 mL) containing 3 mm steel balls and 500 μL buffer solution. Store at ℃ for 10min;
[0086]2) Put the tissue sample into the sample grinder, grind the sample at 25Hz for 1 min, then change the direction, and grind the sample for 1 min again.
[0087] 3) Filter the suspension into a clean centrifuge tube with a 300-mesh nylon mesh, add 4 μL RNase to 120 μL of the filtrate, mix well, and incubate at 37°C for 20-30 minutes;
[0088] 4) Before loading the sample, add 120 μL of staining solution to the mixture and store in the dark.
[0089] (2) Determination and analysis of the ploidy of the samples by using a flow cytometer.
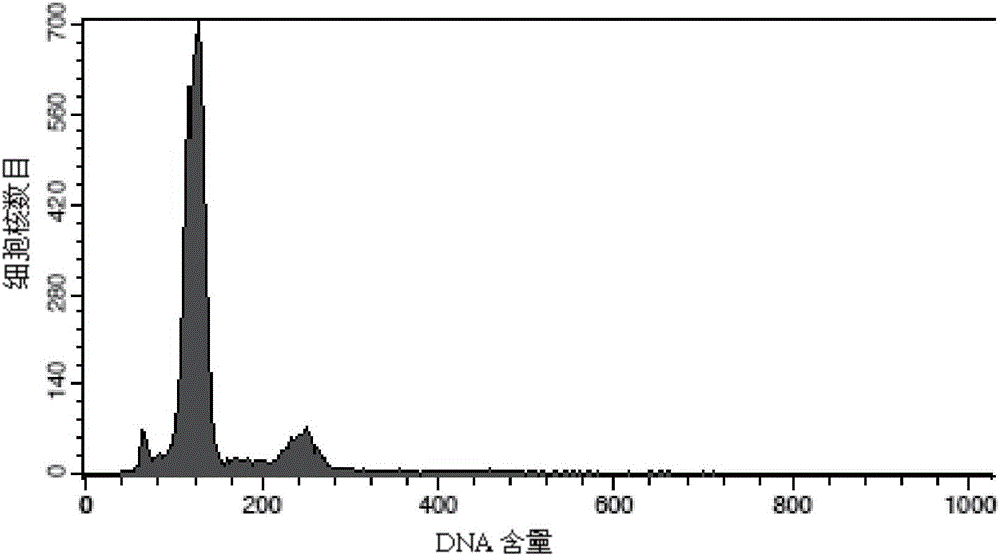
[0090] Test results: The number ...
Embodiment 2
[0092] (1) Treat the tender leaf (from the heart leaf to the third tender leaf) sample with a mill to prepare a single-cell suspension:
[0093] 1) At the four-leaf stage of the plant, take about 50 mg of the third young leaf (the third young leaf from the heart leaf to the outside) and place it in a centrifuge tube (1.5 mL) containing 3 mm steel beads and 500 μL buffer solution. Store at ℃ for 10min;
[0094] 2) Put the tissue sample into the sample grinder, grind the sample at 25Hz for 1 min, then change the direction, and grind the sample for 1 min again.
[0095] 3) Filter the suspension into a clean centrifuge tube with a 300-mesh nylon mesh, add 4 μL RNase to 120 μL of the filtrate, mix well, and incubate at 37°C for 20-30 minutes;
[0096] 4) Before loading the sample, add 120 μL of staining solution to the mixture and store in the dark.
[0097] (2) Determination and analysis of the ploidy of the samples by using a flow cytometer.
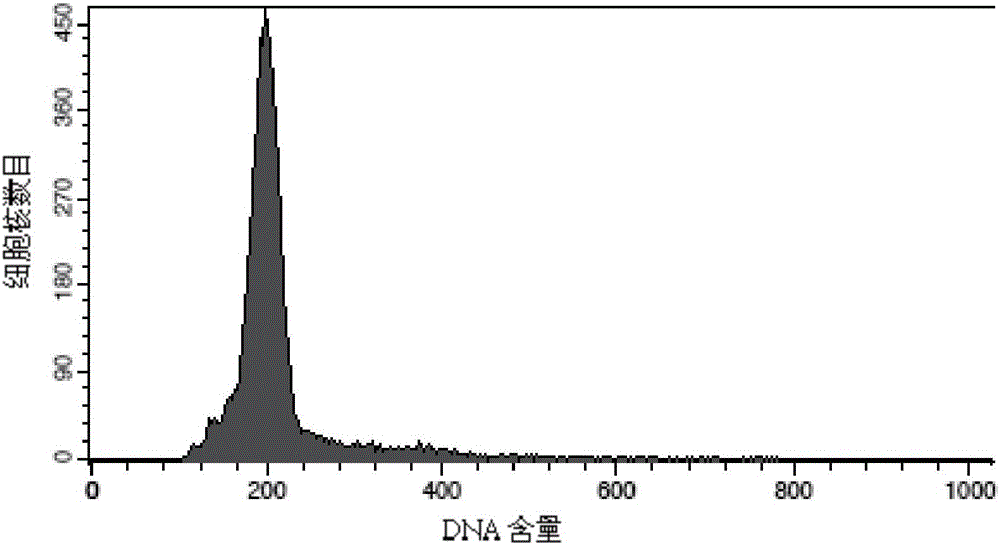
[0098] Test results: The number o...
Embodiment 3
[0100] (1) Treat the tender leaf (from the heart leaf to the third tender leaf) sample with a mill to prepare a single-cell suspension:
[0101] 1) At the five-leaf stage of the plant, take about 50 mg of the third tender leaf (the third tender leaf from the heart leaf to the outside) and place it in a centrifuge tube (1.5 mL) containing 3 mm steel balls and 500 μL buffer solution. Store at ℃ for 10min;
[0102] 2) Put the tissue sample into the sample grinder, grind the sample at 25Hz for 1.5min, then change the direction, and grind the sample for another 1.5min.
[0103] 3) Filter the suspension into a clean centrifuge tube with a 300-mesh nylon mesh, add 4 μL RNase to 120 μL of the filtrate, mix well, and incubate at 37°C for 20-30 minutes;
[0104] 4) Before loading the sample, add 120 μL of staining solution to the mixture and store in the dark.
[0105] (2) Determination and analysis of the ploidy of the samples by using a flow cytometer.
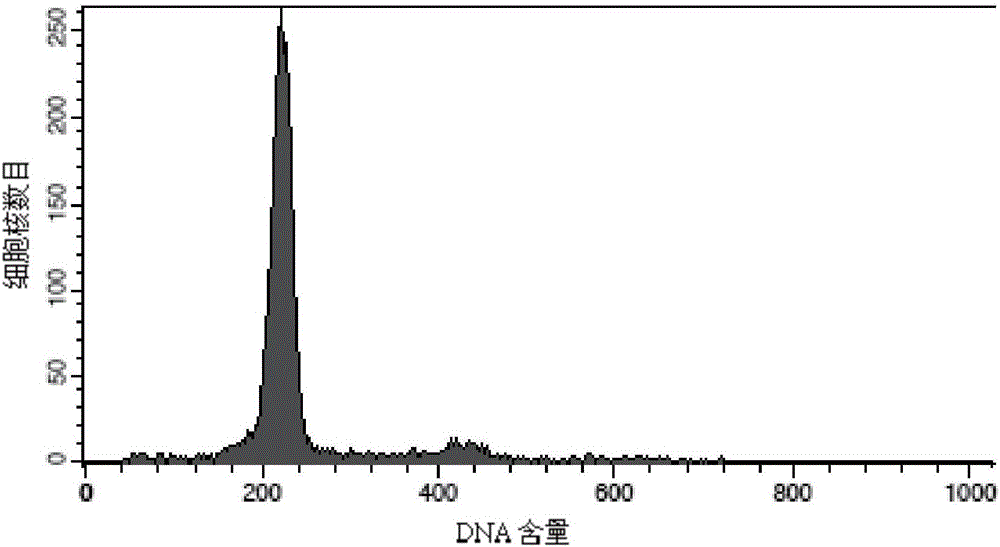
[0106] Test results: The nu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com