CNV detection method and CNV detection apparatus
A technology for sequencing results and genome sequencing, applied in the field of biological information, which can solve the problems of low-depth sequencing data, low genome coverage, and large fluctuations in sequence alignment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: CNV detection method test based on MDA-amplified CG platform low-depth sequencing data
[0039] With the vigorous development of high-throughput sequencing technology, the second-generation sequencing represented by Complete Genomics (CG), IlluminaSolexa and Roche454, as well as the Helicos Genetic Analysis System and single-molecule real-time sequencing technology included in the third-generation sequencing technology (ie, single-molecule sequencing technology) Various sequencing technologies such as (SMRT) and nanopore single-molecule sequencing technology have become important tools for single-cell omics research. As a next-generation sequencing technology focused on the human genome, the CG platform can completely and accurately sequence the entire human genome, and its sequencing throughput is high, which is highly recognized in the industry. The CG platform mainly includes three parts: sequencing platform, high-throughput process automation technology a...
Embodiment 2
[0048] Example 2: CNV detection method test of single-cell low-depth sequencing based on MDA-amplified Proton platform
[0049] Existing single-cell sequencing data are mostly produced by the Illumina sequencing platform. Although the sequencing throughput of the Illumina sequencer is large, its on-machine sequencing time period is long and the sequencing cost is high, which will limit the rapid development of single-cell CNV detection and analysis. However, some studies often have no requirement for sequencing throughput, and relatively have higher requirements for sequencing time and cost. At this time, the Proton sequencing platform will be a better choice. The Proton sequencing platform runs fast, the sequencing cycle only takes a few hours, and the sequencing cost is low. It is more suitable for deployment in hospitals or third-party testing institutions, shortening the testing time, reducing costs, and improving testing efficiency. However, there are few reports on Prot...
Embodiment 3
[0059] Example 3: CNV detection method test of single-cell low-depth sequencing based on MALBAC-amplified Proton platform
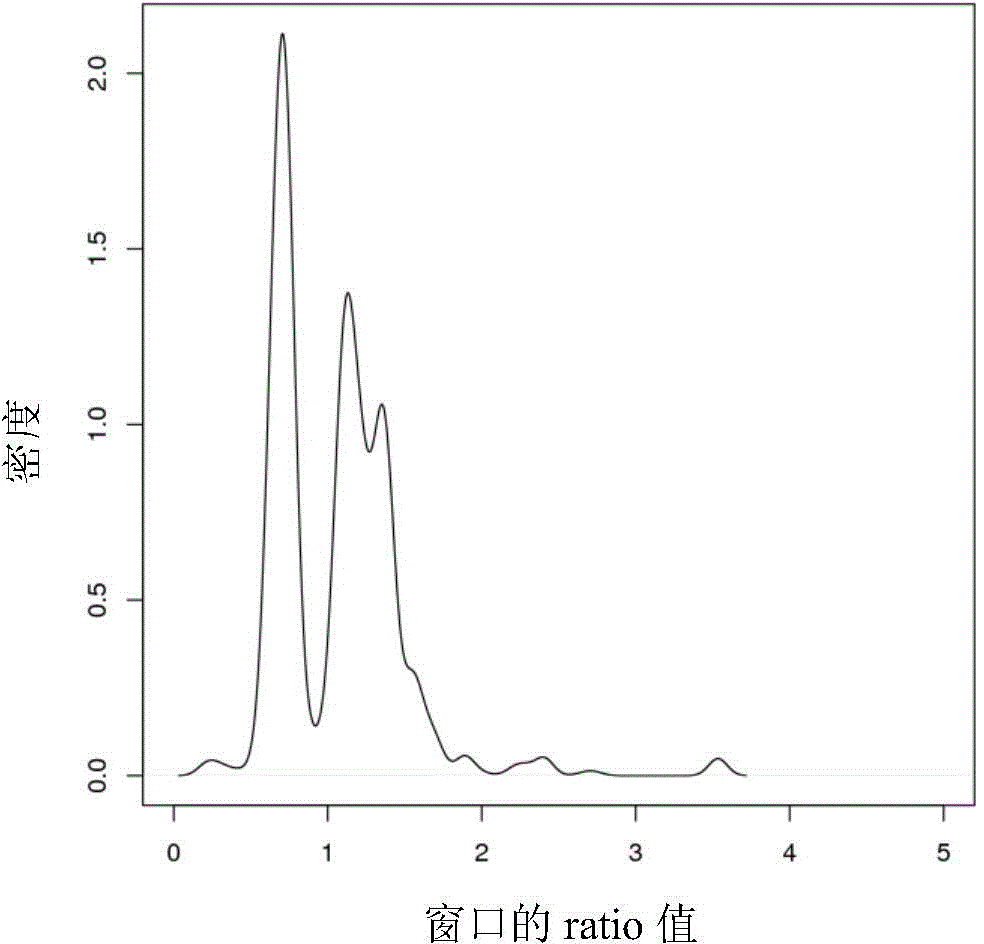
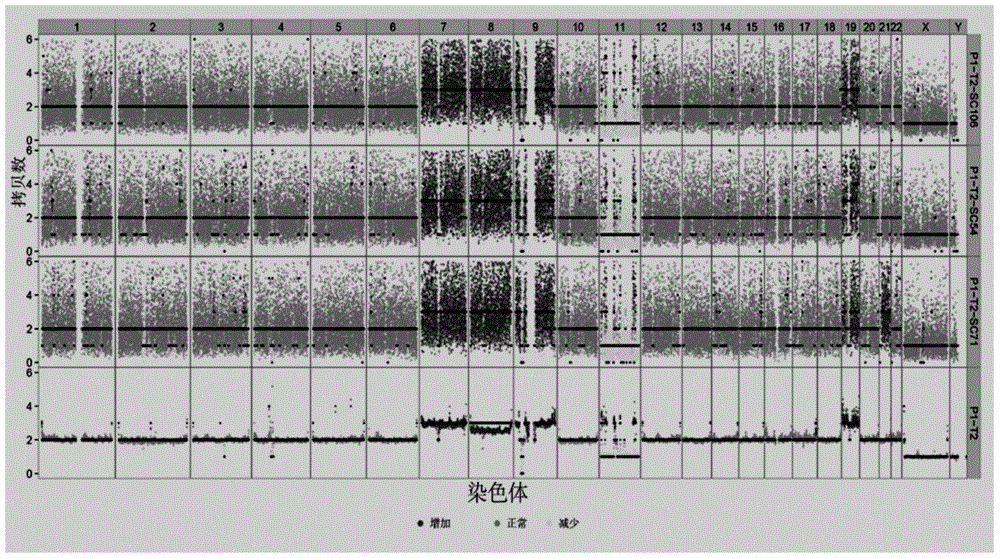
[0060] Five human gastric adenocarcinoma cell lines (BGC823) were extracted, and single-cell low-depth sequencing was performed on the Proton platform after routine library construction by MALBAC whole-genome amplification method; at the same time, a group of cells (BGC ) DNA was sequenced on the Proton platform. The obtained off-board data was tested and analyzed for CNV according to our plan, and CNV was found in five samples of BGC823. The results are as follows Figure 4 As shown, where the abscissa represents the chromosome; the ordinate on the right is 5 single-cell samples and cluster cell samples, and the ordinate on the left is the copy number. The black thick line on the figure represents the ratio value of the divided area, and its value is greater than 2 It means that the copy number in this region is increased, if it is less than 2, it means...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com