Functional marker associated with drought resistance of rape in seedling stage and application of functional marker
A technology for drought resistance and rapeseed seedlings, which is applied in the field of rapeseed breeding and molecular biology, can solve the problems of drought resistance identification, such as heavy workload, long cycle, and easy to be affected by the environment, so as to improve breeding efficiency, facilitate operation, and facilitate popularization and application Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
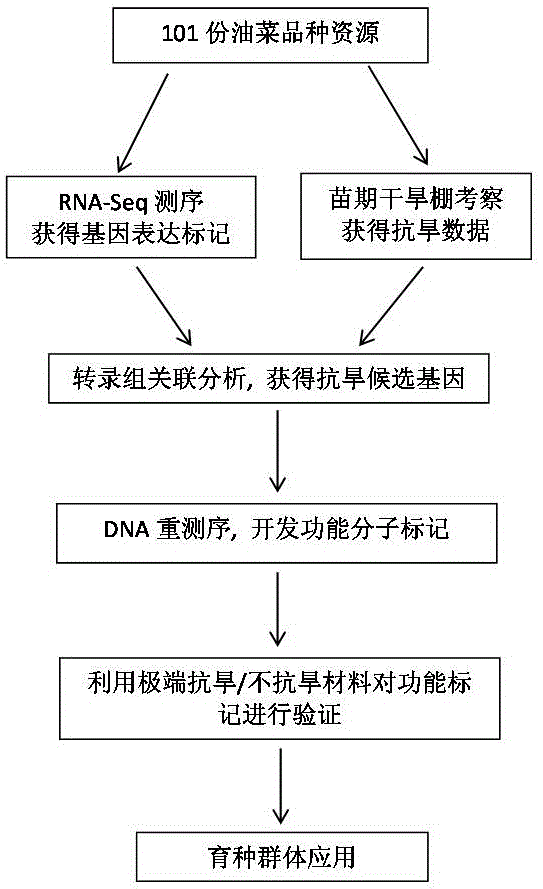
[0033] A preparation method of functional marker EE476451-465 associated with drought resistance in rapeseed seedling stage (Lu Guangyuan et al., Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassicanapus, DNA Research, 2014, 21:613-625), the steps are:
[0034] In this example, taking 101 rapeseed varieties with different genetic sources and large differences in drought resistance as an example, a natural population was constructed, and the method for obtaining molecular markers associated with drought resistance was described in detail, as follows:
[0035] (1) Group construction:
[0036] 101 rapeseed varieties with different genetic origins were used as research materials (published, see literature for details: Lu Guangyuan et al., Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassica napus, DNA Research, 2014, 21(6):613- 625), grown to 1-2 true leaves in a green...
Embodiment 2
[0066] The application of a functional molecular marker highly correlated with drought resistance at seedling stage of rapeseed in drought-resistant breeding comprises the following steps:
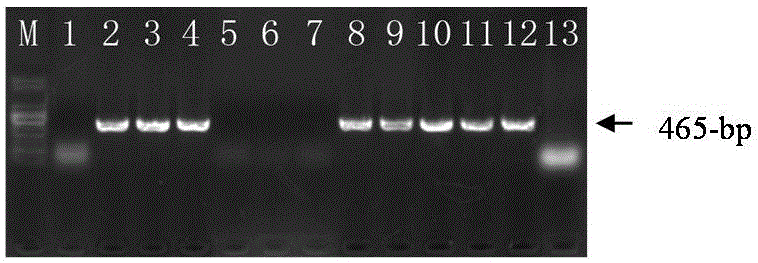
[0067] Select 20 parts of drought-resistant variety materials (the average drought survival rate is 93%) from 101 varieties resources in Example 1, and 20 parts of drought-sensitive materials (the average drought survival rate is 30%), using specific PCR Mark EE476451-465 for identification, the steps are as follows:
[0068] (1) extracting the DNA of the rapeseed strain;
[0069] (2) Perform PCR amplification with functional marker EE476451-465, and the primer sequence is:
[0070] Forward primer EE476451-89F: 5′-TGAGGCTGATAGGATACTGG-3′
[0071] Reverse primer EE476451-554R: 5'-CCACGGGCACCTACATTA-3';
[0072] (3) PCR reaction system: the total volume is 10 μL, and the specific components are as follows:
[0073] DNA template (25ng / μl)
1.0 μL
Forward primer (50ng / μl...
Embodiment 3
[0085] The application of a functional marker highly correlated with drought resistance at seedling stage of rape in drought-resistant breeding of rapeseed comprises the following steps:
[0086] (1) Using the drought-resistant rape variety 'Apex' (derived from Examples 1 and 2) and the poor drought-resistant rape variety 'Zhongshuang 5' (bred by the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, the seeds can be purchased in the market) to construct the F 2 The isolated populations are used as research materials. Samples are taken at the seedling stage, and the detection range is further expanded by using the functional marker EE476451-465. The detection method is the same as that in Example 2.
[0087] (2) It is predicted that the drought resistance of individual plants with 465-bp specific band type (positive) will be significantly improved, while the drought resistance of individual plants without 465-bp specific fragments (negative) will be ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com