SNP marker of medicinal plant murraya paniculata as well as identifying method and application thereof
A technology for medicinal plants and gulira, which is applied in the biological field to achieve the effects of simple identification process, stability and reliability, and wide applicability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
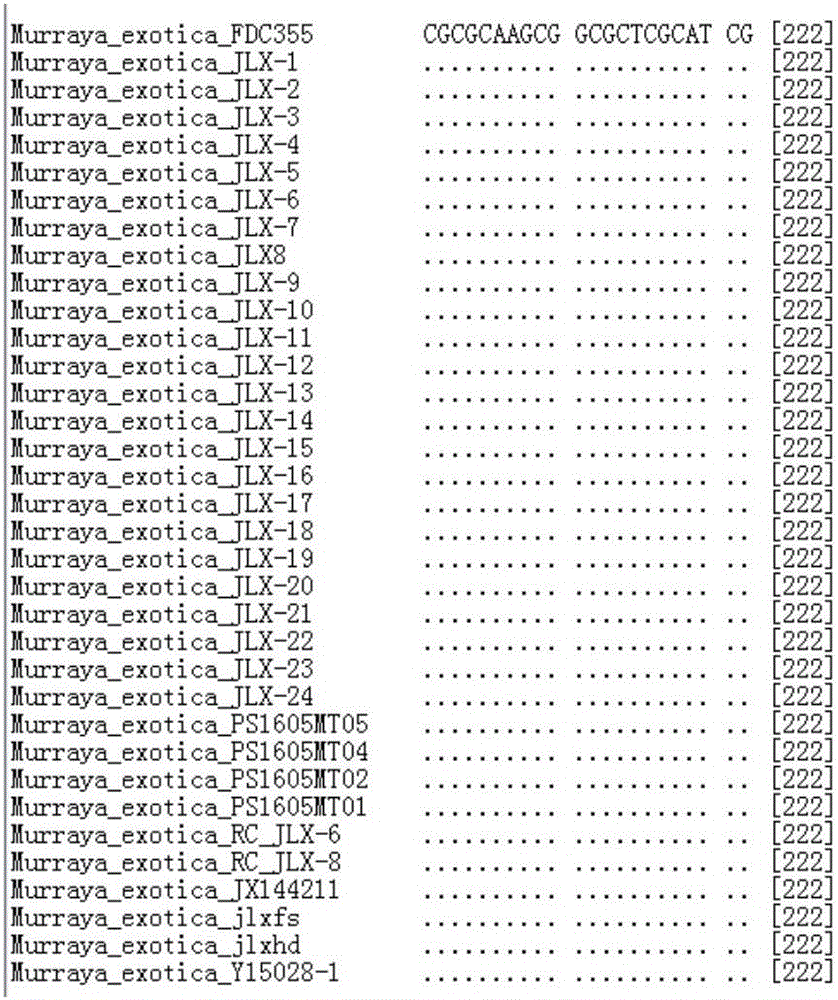
Image
Examples
Embodiment 1
[0049] The rapid molecular identification method of the medicinal plant Murata chinensis described in the present embodiment comprises the following steps:
[0050] S1. Extract the total DNA of each of the samples of Murata to be tested according to conventional methods, and the extraction steps are as follows: take the leaves (about 30 mg) of the samples of Murata to be tested respectively, and grind them on a MM400 ball mill, Then use the Plant Genomic DNA Extraction Kit (TiangenBiotechCo., China) to extract the total genomic DNA for subsequent use;
[0051] S2. Using the total genomic DNA obtained in step S1 as a template, design the following primers:
[0052] ITS2F:5'-ATGCGATACTTGGTGTGAAT-3',
[0053] ITS3R:5'-GACGCTTCTCCAGACTACAAT-3', said primers are shown in SEQIDNo.2 and SEQIDNo.3, carry out PCR amplification to described total genomic DNA, and described PCR amplification reaction system comprises:
[0054] DNA template, 50ng, 2μL;
[0055] Forward and reverse prim...
Embodiment 2
[0061] The rapid molecular identification method of the medicinal plant Murata chinensis described in the present embodiment comprises the following steps:
[0062] S1. According to the conventional method, extract the total DNA of the medicinal Murata to be tested and the samples of the closely related species of Murata. The extraction steps are as follows: take about 30 mg of the leaves of the samples to be tested respectively, and conduct them on the MM400 ball mill. Grind, then extract the total genomic DNA with a plant genomic DNA extraction kit (TiangenBiotechCo., China), and set aside;
[0063] S2. Using the total genomic DNA obtained in step S1 as a template, design the following primers:
[0064] ITS2F:5'-ATGCGATACTTGGTGTGAAT-3',
[0065] ITS3R: 5'-GACGCTTCTCCAGACTACAAT-3', performing PCR amplification on the total genomic DNA, the PCR amplification reaction system comprising:
[0066] DNA template, 50ng, 2μL;
[0067] Forward and reverse primers, 2.5 μM, 1 μL each...
Embodiment 3
[0073] The rapid molecular identification method of the medicinal plant Murata chinensis described in the present embodiment comprises the following steps:
[0074] S1. Extract the total DNA of each sample of the medicinal Murata to be tested according to a conventional method. The extraction steps are as follows: take 1 seed of the sample of Murata to be tested, grind it on a MM400 ball mill, and then use Plant Genomic DNA Extraction Kit (TiangenBiotechCo., China) was used to extract the total genomic DNA and set aside;
[0075] S2. Using the total genomic DNA obtained in step S1 as a template, design the following primers:
[0076] ITS2F:5'-ATGCGATACTTGGTGTGAAT-3',
[0077] ITS3R:5'-GACGCTTCTCCAGACTACAAT-3', said primers are shown in SEQIDNo.2 and SEQIDNo.3, carry out PCR amplification to described total genomic DNA, and described PCR amplification reaction system comprises:
[0078] DNA template, 40ng, 2μL;
[0079] Forward and reverse primers, 2.5 μM, 1 μL each;
[008...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com