Fusion peptide TAT-MAS9C for destroying interaction between Mas receptors and PSD95
A TAT-MAS9C, fusion peptide technology, applied in the field of fusion peptides, can solve the problems of inappropriate PSD95 regulation function research, low neuronal transfection efficiency, etc., and achieve the effect of strengthening the protective effect of cerebral ischemia
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Design and synthesis of the fusion peptide of the present invention
[0032] The fusion peptide TAT-MAS9C used to destroy the interaction between the Mas receptor and PSD95 has a full length of 20 amino acids, and its sequence includes two parts. The amino terminal of the fusion peptide is the transmembrane domain of the HIV-1 Tat protein. The 11 amino acids YGRKKRRQRRR can introduce the fusion peptide into the cytoplasm; the carboxy-terminal of the fusion peptide is the 9 amino acids NTVSIETVV at the carboxy-terminal of Mas, which can destroy the interaction between Mas and PSD95. The amino acid sequence of the fusion peptide is YGRKKRRQRRRNTVSIETVV.
[0033] For some research needs, rhodamine can be attached to amino acid K of the fusion peptide for fluorescent labeling. The method for producing the fusion peptide is well known, and general peptide synthesis companies can synthesize it by solid-phase synthesis.
[0034] The method for synthesizing the fu...
Embodiment 2
[0036] Example 2 Verification of the transmembrane effect of TAT-MAS9C by fluorescence microscopy
[0037] Program:
[0038] The experiment was divided into two groups: group A was rhodamine-labeled negative control group; group B was TAT-MAS9C group. Primary cultured neurons for 7 days, after the synapse grows, use rhodamine-labeled negative control and 10 -5 M's TAT-MAS9C incubated neurons for 30 minutes, washed away the negative control and TAT-MAS9C, and took pictures with a fluorescence microscope to observe their transmembrane effects.
[0039] result:
[0040] After washing off the peptide, it was found that the negative control in group A had a poor penetration effect and the fluorescence intensity was very weak ( Figure 1A ). In the cytoplasm of neurons in group B, a large number of fluorescently labeled TAT-MAS9C ( Figure 1B ), indicating that TAT-MAS9C has a good transmembrane effect.
Embodiment 3
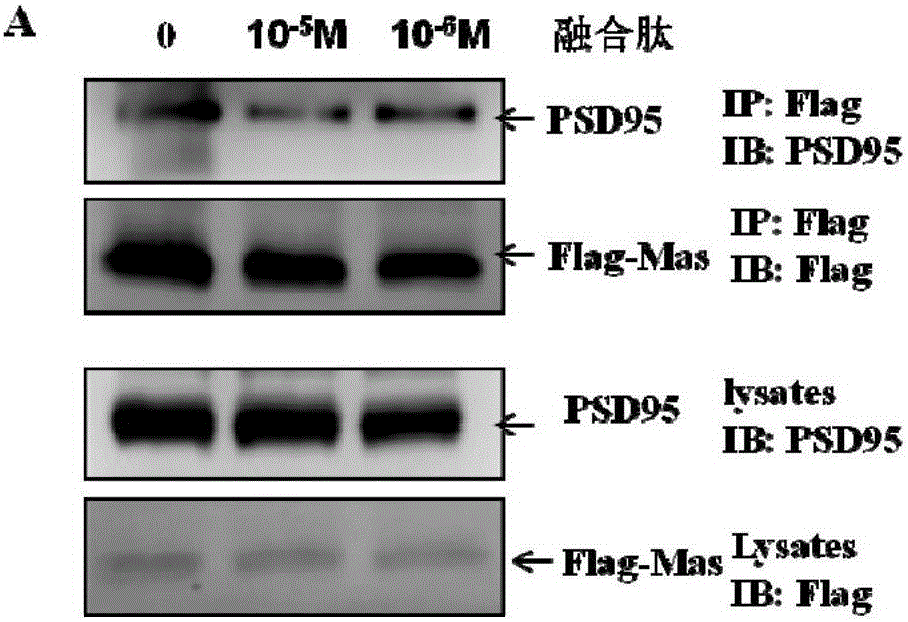
[0041] Example 3 The interference effect and specificity of TAT-MAS9C were verified by CoIP (co-immunoprecipitation) combined with western blot (immuno-western blotting) method:
[0042] Program:
[0043] The experiment was divided into three groups. The 293 cells were seeded in 90mm cell culture dishes, and when the density was about 70%, the 293 cells were transfected with Flag-Mas and PSD95 plasmids at the same time. 24 hours after transfection, add the negative control to the culture medium of group A and incubate for 30 minutes; add the final concentration of 10 -5 M's TAT-MAS9C, incubated for 30min; C group added 10 -6 M TAT-MAS9C, incubated for 30min. Cells were harvested with protein lysate, mixed continuously at 4°C for 1 h, and the supernatant was collected by centrifugation. Add 50 μL of Anti-FlagM2AffinityGel to 1 mL of supernatant, and keep mixing for 3 hours at 4°C. Centrifuge for 1 min to collect the precipitate, wash the precipitate with washing solution,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com