Application of a Molecular Marker in Association Analysis of Milk Yield and Protein Content Traits in Dairy Cows
A technology of protein content and molecular markers, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as not identifying SNPs, achieve shortened generation intervals, low cost, and intuitive pictures Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Implementation example 1: application example
[0038] An application of a molecular marker MAP4K4 fragment in the correlation analysis of milk yield and protein content traits of dairy cows, the application steps are as follows:
[0039] 1) Primer design:
[0040] According to the partial sequence of the 18th exon of the MAP4K4 gene in the NCBI website (GenBank sequence number AC_000168.1), the applicant designed amplification primers containing the partial sequence, and the primer synthesis was completed by Shanghai Bioengineering Technology Service Co., Ltd. The DNA sequence is as follows:
[0041] P1 forward primer: 5'GGCTCCACTTCACAATACC 3',
[0042] P2 reverse primer: 5'TCTGACTCTGTGCAACCC 3'.
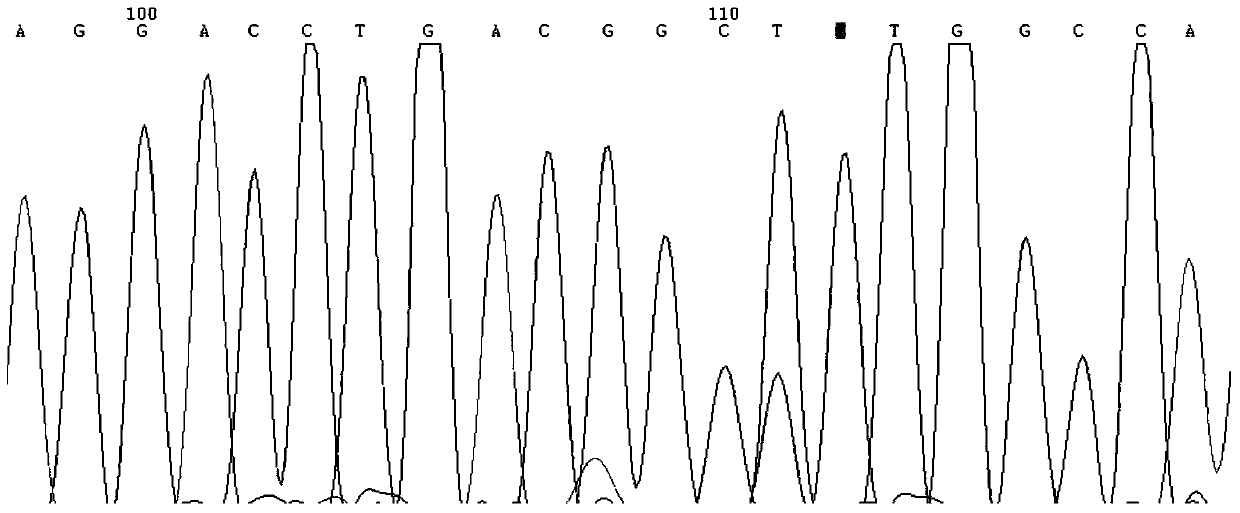
[0043] 2) Purification and sequencing of PCR products:
[0044] PCR amplification reaction system 20ul, including 500ng DNA, 2.0ul 10×PCR buffer, 0.6ul 10mMdNTP, 0.8ul of each primer (10uM) and 1U Ampli Taq DNA polymerase (Shanghai Sangon Bioengineering Technology Servic...
Embodiment 2
[0051] Implementation example 2: association analysis of traits such as dairy cow milk production and protein content related to MAP4K4 gene polymorphism site T156-G156
[0052] After detecting the genotypes of the molecular marker T156-G156 in 413 Holstein dairy cows by PCR-RFLP, statistical methods were used to analyze the correlation between the loci and the milk production and protein content of the cows. Statistical analysis uses R language, using the least multiplication to fit the linear model, the formula is as follows:
[0053] Y ijk =u+F i +P j +SNP k +e ijk
[0054] where Y ijk is the phenotype value, u is the population mean, F i is the site effect results show that P j is a sex effect, the SNP k is the SNP effect, e ijk is a random error.
[0055] Correlation analysis showed that the protein content of individuals with TT genotype was significantly higher (P=0.015) than that of GG genotype, and the milk production of individuals with GG genotype was sig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com