Method for automatically and quickly judging fluorescent quantitative PCR (Polymerase Chain Reaction) result
A fluorescent quantitative and rapid technology, applied in the field of biological analysis, can solve the problems of inability to analyze, consume a lot of time, reduce experimental efficiency, etc., and achieve the effect of improving judgment speed and accuracy, improving recognition ability, and reducing false positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The present invention will be further described in detail below in conjunction with the accompanying drawings, so that those skilled in the art can implement it with reference to the description.
[0026] This case lists an embodiment of a method for automatically and rapidly discriminating fluorescent quantitative PCR results, which includes:
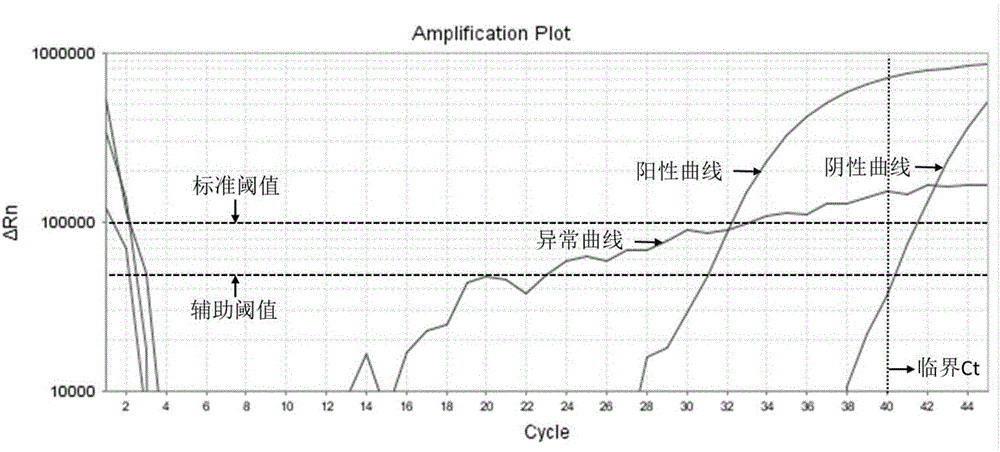
[0027] Extract the data curve of fluorescent quantitative PCR;
[0028] Baseline the data curve;
[0029] Set up a standard threshold value of fluorescence intensity, the corresponding cycle number is Ct1, and the standard threshold value is set in the exponential rising period of the data curve;
[0030] Set up an auxiliary threshold value of fluorescence intensity, the corresponding cycle number is Ct2, the auxiliary threshold value is set in the exponential rising period of the data curve, and the standard threshold value≠auxiliary threshold value;
[0031] Set up a scoring formula z to calculate the score, scoring formula ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com