A Rapid and Accurate Method for Identifying Contamination Sources in High-Throughput Genomic Data
A high-throughput, genomic technology, applied in the field of molecular biology, which can solve the problems of data analysis impact, inevitability, and inaccurate evaluation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
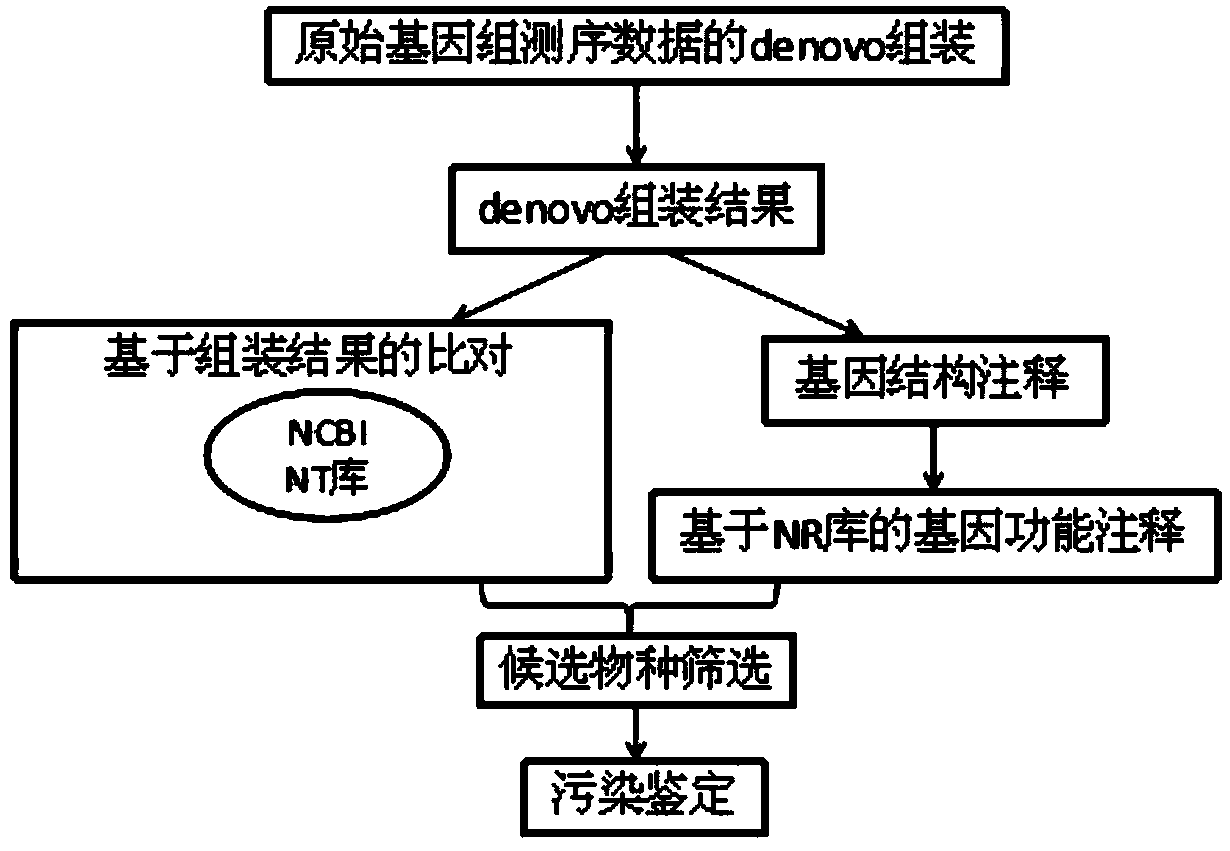
[0037] Denovo sequencing of the genome of a pathogenic fungus (Plasmopara halstedii), the second-generation illumina platform has two libraries of 180bp and 500bp, the sequencing depths are 35X and 34X, and the length of each read is 100bp. The total number of reads in each library is respectively For 46308070 and 43435185, a total of 89743255 items, with a total data volume of 8.36G, use the following methods to identify pollution sources:
[0038] (1) Assemble using ABYSS software (k-mer parameter is set to k=50, other parameters are software default parameters), the number of scaffolds in the assembly result is 30428 in total, N50 is 10506, the longest is 479848, and the size is 80M; you can It is easy to see that: ①The total number of assembled sequences is 30428, which is only 0.03% of the original total number of 89743255 sequences; ②The total data volume is 118M, which is only 1.38% of the original 8.36G total data volume. ③The sequence length is increased from 100bp to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com