Set of genes for head and neck squamous cell carcinoma (HNSCC) molecular typing and application thereof
A squamous cell carcinoma and molecular typing technology, which is applied in the field of preparation of kits for molecular typing of head and neck squamous cell carcinoma, can solve the problem of inability to guide HNSCC personalized treatment, limited research, and inaccurate evaluation methods Advanced problems to achieve the effect of improving the ability of clinical treatment, strengthening the rationality and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Screening to Assess HNSCC Molecularly Typing Genomes
[0023] Through the EPIG gene expression profiling analysis program, 789 cases of HNSCC tumor gene whole gene sequencing, exome sequencing, RNA sequencing, gene expression chip and clinical variable related data that have been published or stored in databases such as TCGA and COSMIC were analyzed to screen and evaluate HNSCC molecules Typing Genomes. Analysis included unsupervised cluster analysis to identify expression profiles and associated genes; stability analysis of candidate genes. Firstly, the correlation coefficient of each gene in 789 samples was calculated by cluster analysis and clustered to form a gene-specific expression profile (r>0.7, P0.7, P<0.001). Experimental results: The 18 genes with the highest scores for evaluating the molecular typing of HNSCC were screened, and the gene list is shown in Table 1 below.
[0024] Table 1: 18 genes that can be used to assess molecular typing of HNSC...
Embodiment 2
[0026] Example 2 Molecular typing of HNSCC assessed by digital PCR method and gene expression microarray
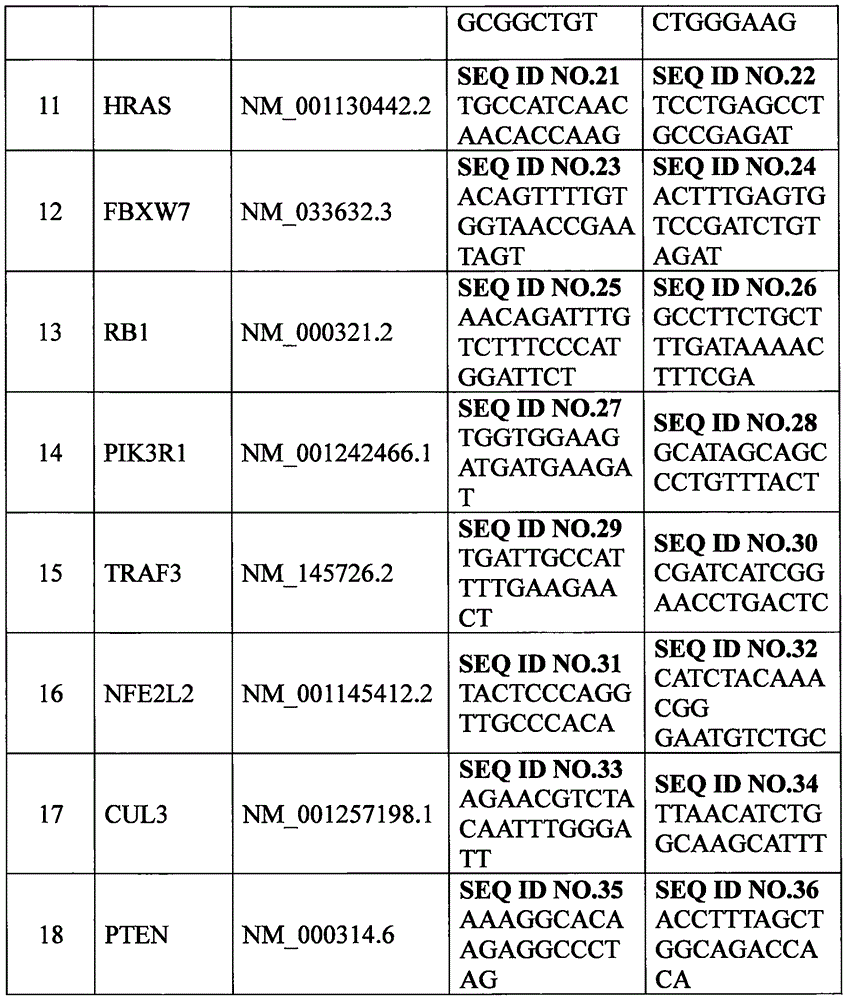
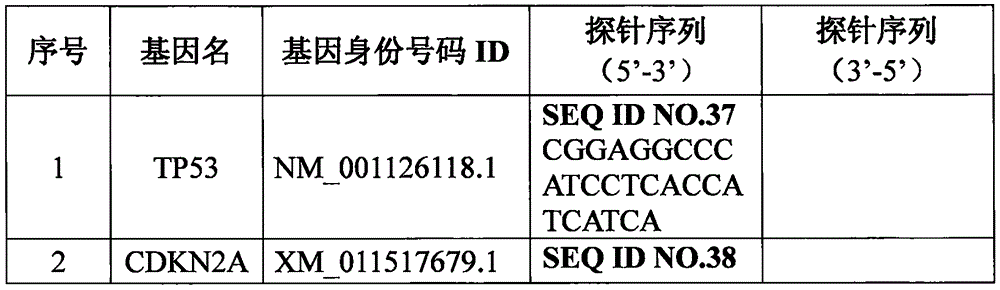
[0027] In this example, digital PCR and gene expression chip methods were used to detect the expression levels of 18 genes screened in Example 1 in paraffin-embedded tumor tissues and their matched paracancerous tissues of 100 groups of Chinese HNSCC patients. The primers and probes used in digital PCR detection were designed according to the complete human gene sequence released by NCBI (National Center for Biotechnology Information), using Primer Premier 5.0, and synthesized by Gene Synthesis Company (Shanghai Sangong). The nucleotide sequences of the primers are shown in Table 2 below (SEQ ID No. 1 to SEQ ID No. 36 in the sequence listing), and the nucleotide sequences of the probes are shown in Table 3 below (SEQ ID No. 37 to SEQ ID No. 54 in the sequence listing.
[0028] Table 2: PCR primer sequences for 18 genes that can be used to assess molecular typing of HNSC...
Embodiment 3
[0039] Example 3 Molecular typing of HNSCC assessed by gene expression microarray
[0040] In this example, the gene expression microarray method was used to detect the expression levels of the 18 genes screened in Example 1 in the same 100 groups of Chinese HNSCC patient paraffin-embedded tumor tissues and their matched paracancerous tissues as in Example 2 . The probes used in the gene expression chip were designed according to the human whole gene sequence published by NCBI (National Center for Biotechnology Information), using Primer Premier 5.0, and synthesized by Gene Synthesis Company (Shanghai Sangong). The nucleotide sequences of the probes are shown in SEQ ID No. 37 to SEQ ID No. 54 in the sequence listing.
[0041] The gene expression chip is to immobilize a large number of DNA fragments or oligonucleotide fragments of specific sequences on glass, silicon wafers and other carriers at high density in a matrix through microelectronic technology and microprocessing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com