Use of Ell as an E3 ubiquitin ligase
A sequence and protein technology, applied in the application field of ELL as E3 ubiquitin ligase, can solve the problems of non-expression and inactivation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
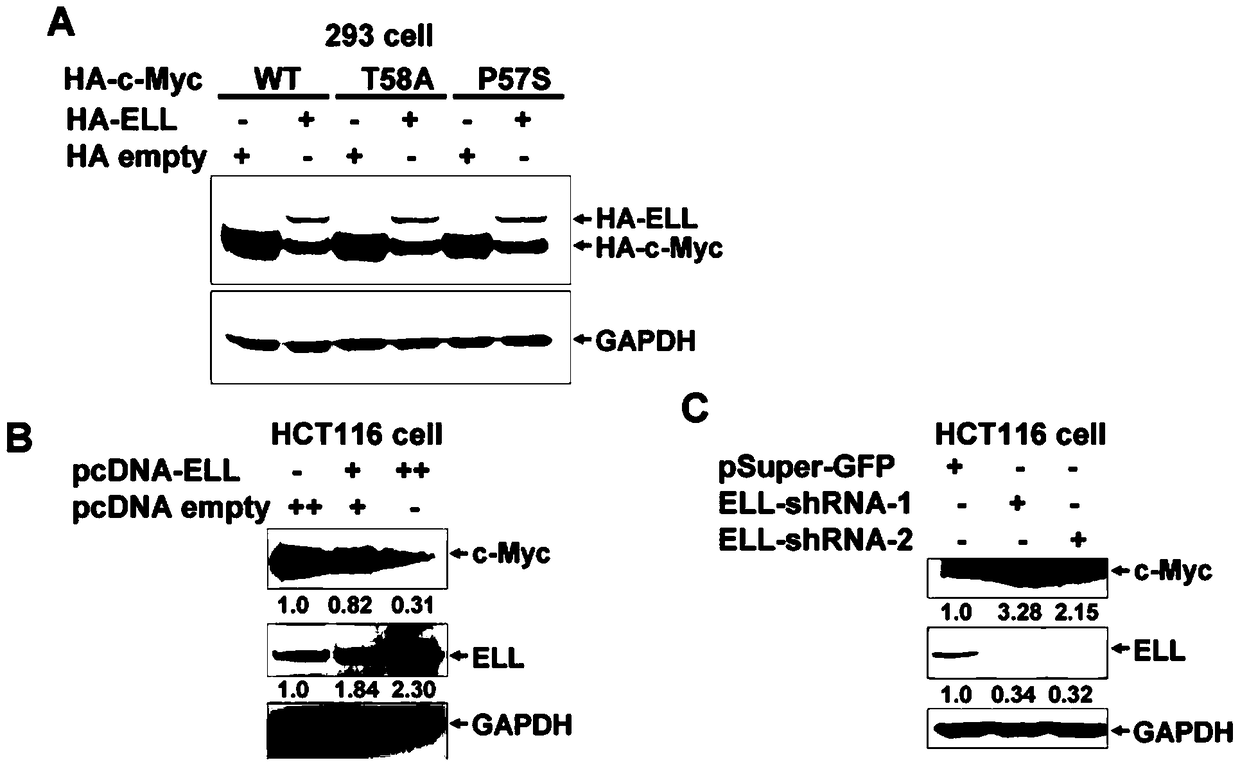
[0077] Example 1, ELL promotes c-Myc degradation
[0078] The amino acid sequence of protein c-Myc is sequence 2, and the nucleotide sequence of its coding gene is sequence 1;
[0079] The amino acid sequence of protein ELL is sequence 4, and the nucleotide sequence of its coding gene is sequence 3.
[0080] The above sequence was artificially synthesized.
[0081] 1. ELL reduces the expression of exogenous Myc
[0082] The plasmid HA-c-Myc (WT) expressing c-Myc is a plasmid obtained by inserting the c-Myc coding gene into the XbaI and BamHI sites of the pCMV-HA vector (Clontech);
[0083] The plasmid HA-ELL expressing ELL is a plasmid obtained by inserting the coding gene of ELL into the SalI and KpnI sites of the pCMV-HA vector;
[0084] Plasmid HA-empty is the pCMV-HA vector.
[0085] Plasmids HA-ELL and HA-empty were co-transfected with HA-c-Myc(WT) at a mass ratio of 1:1 to HEK293 cells. On the second day after transfection, the supernatant was collected for Western ...
Embodiment 2
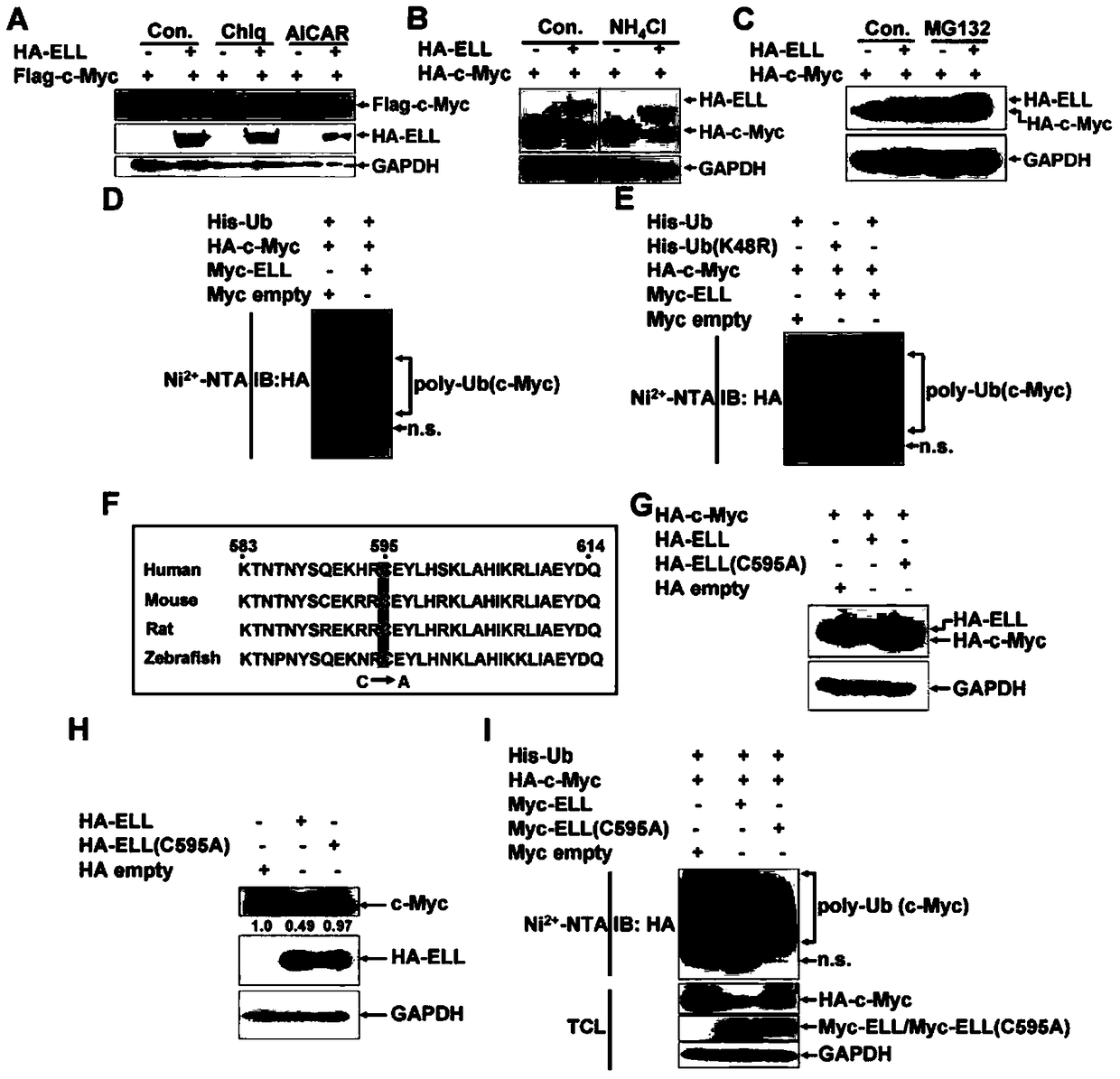
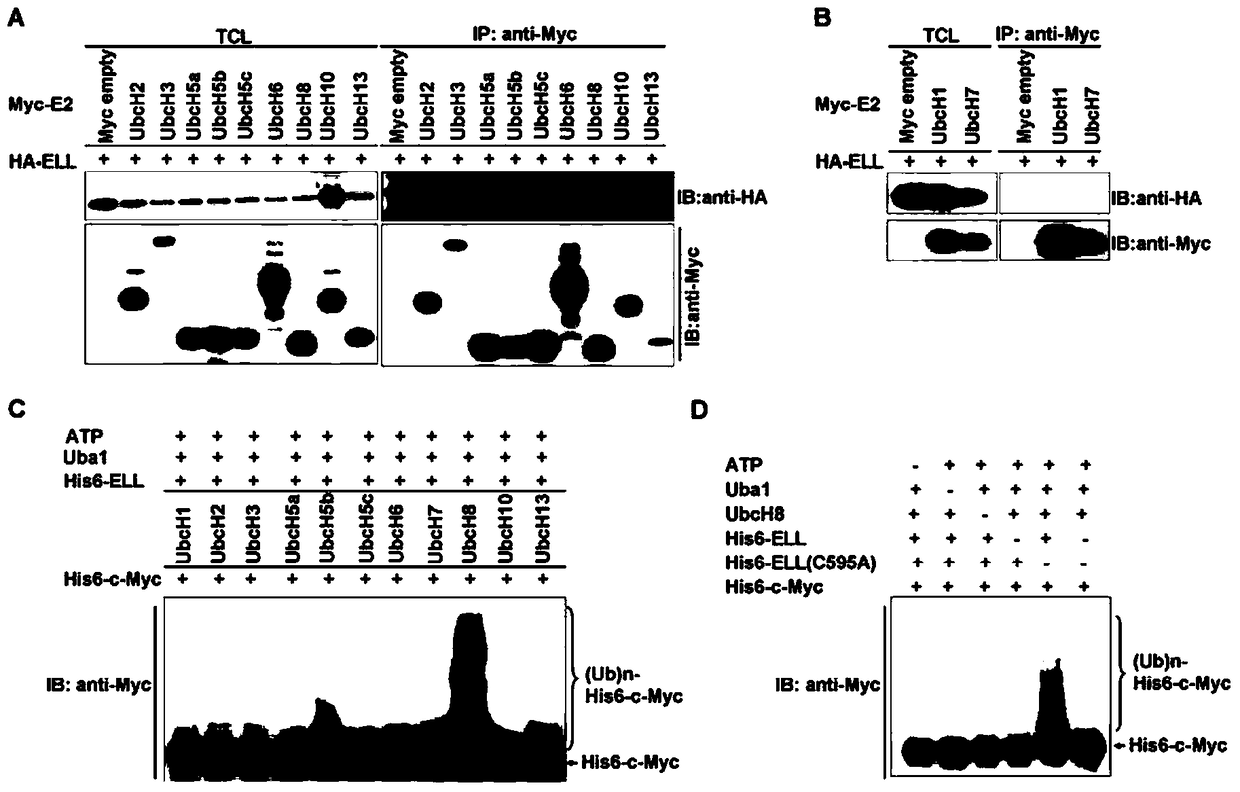
[0102] Example 2, ELL acts as an E3 ubiquitin ligase to degrade c-Myc protein
[0103] 1. ELL can enhance the ubiquitination of c-Myc
[0104] In order to verify the type of protein degradation caused by ELL, inhibitors were used, including chloroquine (lysosomal proteolysis inhibitor, Chlq) (Sigma, 87111906), ammonium chloride (lysosomal proteolysis inhibitor, NH 4 CL) (Sinopharm, 12125-02-9), AICAR (autophagy inhibitor) (Cayman, 2627-69-2) and MG132 (proteasome inhibitor) (Sigma, M8699) to determine whether they can block ELL-mediated degradation of c-Myc.
[0105] Plasmid HA-ELL, plasmid HA-ELL and plasmid HA-c-Myc were transfected into HEK293 cells respectively. On the second day after transfection, Chlq and AICAR were added to the cell culture medium respectively, and the final concentrations of Chlq and AICAR reached 20mM, continue to culture for 6 hours, collect the supernatant for Western blot detection, the antibody used is shown in the figure. Take no inhibitor as...
Embodiment 3
[0175] Example 3, ELL inhibits the tumor growth of colon xenografts
[0176] The HT116 cells transfected with pHAGE-control, the HT116 cells transfected with pHAGE-ELL, and the HT116 cells transfected with pHAGE-ELL (C595A) in Example 2 were subjected to transplantation tumor growth experiments: 15 male nude mice were randomly divided into 3 groups , 5 in each group, three groups of nude mice were subcutaneously injected with the above three different cell lines, and the amount of cells injected into each nude mouse was 2×10 6 indivual. From the third week, the length, width, and height of the tumor were measured weekly and the tumor volume was calculated using V=π.abc / 6 (Flatz et al. 2010). After 6 weeks, the nude mice were sacrificed, the weight of the tumor was measured and the corresponding gene expression was analyzed. Lung tissues were analyzed histologically after HE staining.
[0177] Tumor volume results such as Image 6 A and 6B, the tumor growth rate of HCT116 c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com