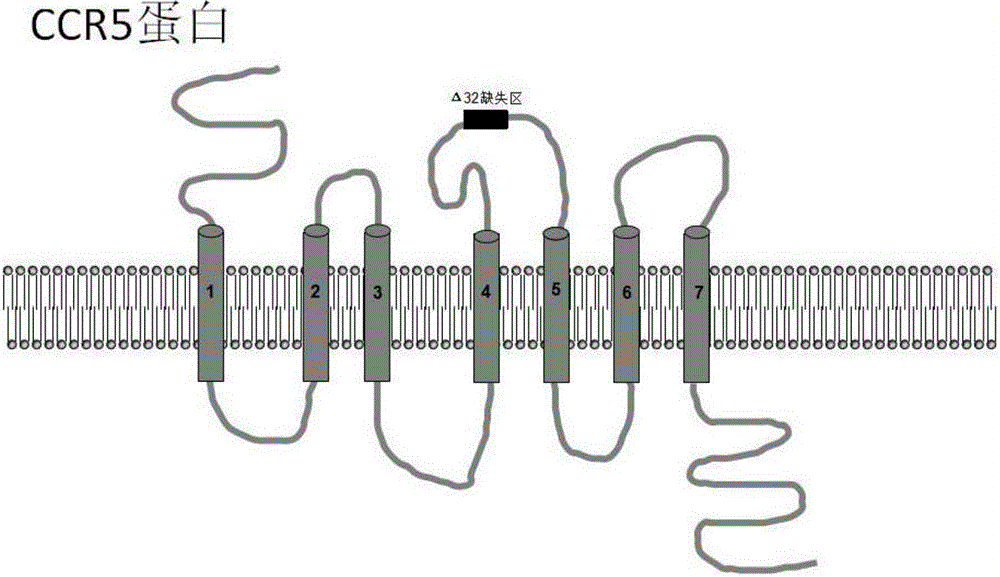
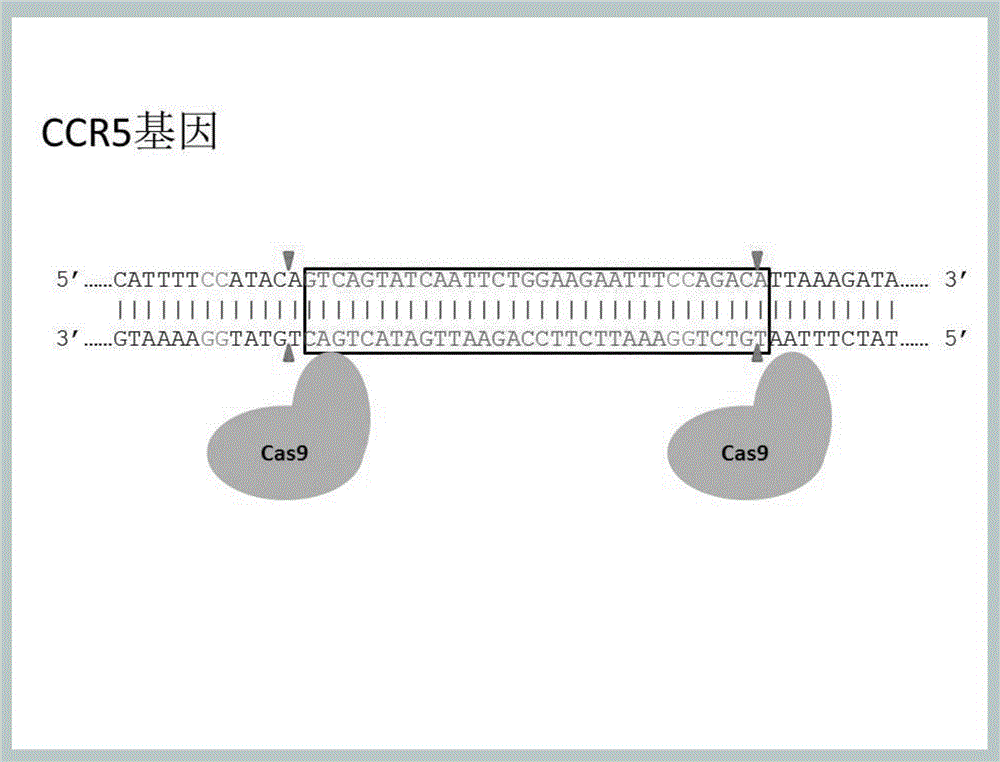
Method for inducing CCR5-delta32 deletion with genome editing technology CRISPR-Cas9
A genome editing and technology technology, applied in recombinant DNA technology, gene therapy, polymorphism use, etc., can solve problems such as CCR5Δ32 deletion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Plasmid Reconstruction Carrying CRISPR-Cas9 System and Preparation of Lentiviral Particles Encapsulating the Reconstructed Plasmid
[0038] 1. Selection and design of gRNA
[0039] (1) According to the purpose of the experiment, we need to design a pair of gRNAs to edit and modify the CCR5 gene at the same time. However, due to the limitations of the CCR5 gene and the limitations of the PAM site, the choice of gRNA is limited. The left gRNA, such as SEQIDNo.1 , or SEQNo.3-19. The right sequence, such as SEQ ID No.2 or SEQ ID No.20-36. Or any combination of these flanking gRNAs.
[0040] (2) According to the gRNA sequence, enzyme cleavage sites were added to synthesize DNA, which were respectively connected into the lenti-CRISPR-v2 vector digested by BsmBI to obtain the reconstructed plasmid, and the correctness of the sequence was verified by sequencing. The whole experimental process, such as Figure 4 .
[0041] 2. Preparation of lentiviral particles a...
Embodiment 2
[0044] Example 2: Results of Lymphocytes Infected with Lentiviral Particles Encapsulated with the Reconstituted Lenti-CRISPR-v2 Plasmid
[0045] 1. Spread lymphocytes in a six-well plate before infection, and the total number of cells in each well is 2×10 5 , the medium used is a full medium without antibiotics. On the second day, the lentiviral particles of the lenti-CRISPR-v2 plasmid wrapped with gRNA obtained in Example 1 and the lentiviral particles of the empty lenti-CRISPR-v2 plasmid wrapped The disease particles were added to the cell culture medium, and polybrene, which can increase the efficiency of virus infection, was added at the same time. Culture medium, after 72 hours of culture, the cells were collected by centrifugation, and half of the collected cells were transferred to a new culture flask to continue culturing, and the other half was used to extract the total genomic DNA of the cells for subsequent analysis.
[0046] 2. Using the extracted cellular genomic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com