Method for detecting quarantine Xanthomonas and special primers thereof
A Xanthomonas and primer pair technology, applied in the field of microbiology, can solve the problems of not meeting the requirements of rapid customs clearance for plant quarantine at ports, time-consuming, complicated operations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Embodiment 1, design and synthesis of primers
[0072] The type III secretion system of pathogenic bacteria plays a crucial role in the interaction between pathogens and hosts. The hrp (hypersensitive response & pathogenicity) gene encoding type III secretion system widely exists in Gram-negative plant pathogenic bacteria, and its gene cluster is composed of hrc (hypersensitive response & conserved), hpa (hypersensitive response & associated), hrp, and some transposons. Through preliminary experiments and sequence comparison, it was found that the hpaA gene sequence is relatively conservative within the species, but has certain variability among species, which is a good tool for distinguishing and identifying different species and pathogenic variants of Xanthomonas barcoded genes.
[0073] According to the hpaA gene of Xanthomonas quarantineis, two primer pairs were designed, named primer pair A and primer pair B respectively. The primer pair A is composed of primer X...
Embodiment 2
[0080] Embodiment 2, establishment of the method for detecting quarantine Xanthomonas
[0081] The established method is as follows:
[0082] 1. Extract the genomic DNA of the bacterium to be tested and use it as a template to carry out PCR amplification using the synthetic primer pair A or primer pair B in Example 1 to obtain a PCR amplification product.
[0083] Reaction system: Genomic DNA (6—50) ng of the sample to be tested, Taq PCR Mastermix (product of Tiangen Biochemical Technology (Beijing) Co., Ltd., catalog number KT201-02) 12.5 μL, primer pair A (primer XHpaAT-F and primer XHpaAT-R each 10 pmol) or primer pair B (primer XHpaAH-F and primer XHpaAH-R each 10 pmol), make up to 25 μL with deionized water.
[0084] Reaction program: 94°C for 5min; 94°C for 40S, 55°C for 40S, 72°C for 1min, 35 cycles; 72°C for 10min.
[0085] 2. After step 1 is completed, the PCR amplification product is sequenced, and the following judgment is made according to the sequencing result: ...
Embodiment 3
[0098] Embodiment 3, identify whether 58 kinds of test bacterial strains are quarantine Xanthomonas according to the method established in Example 2
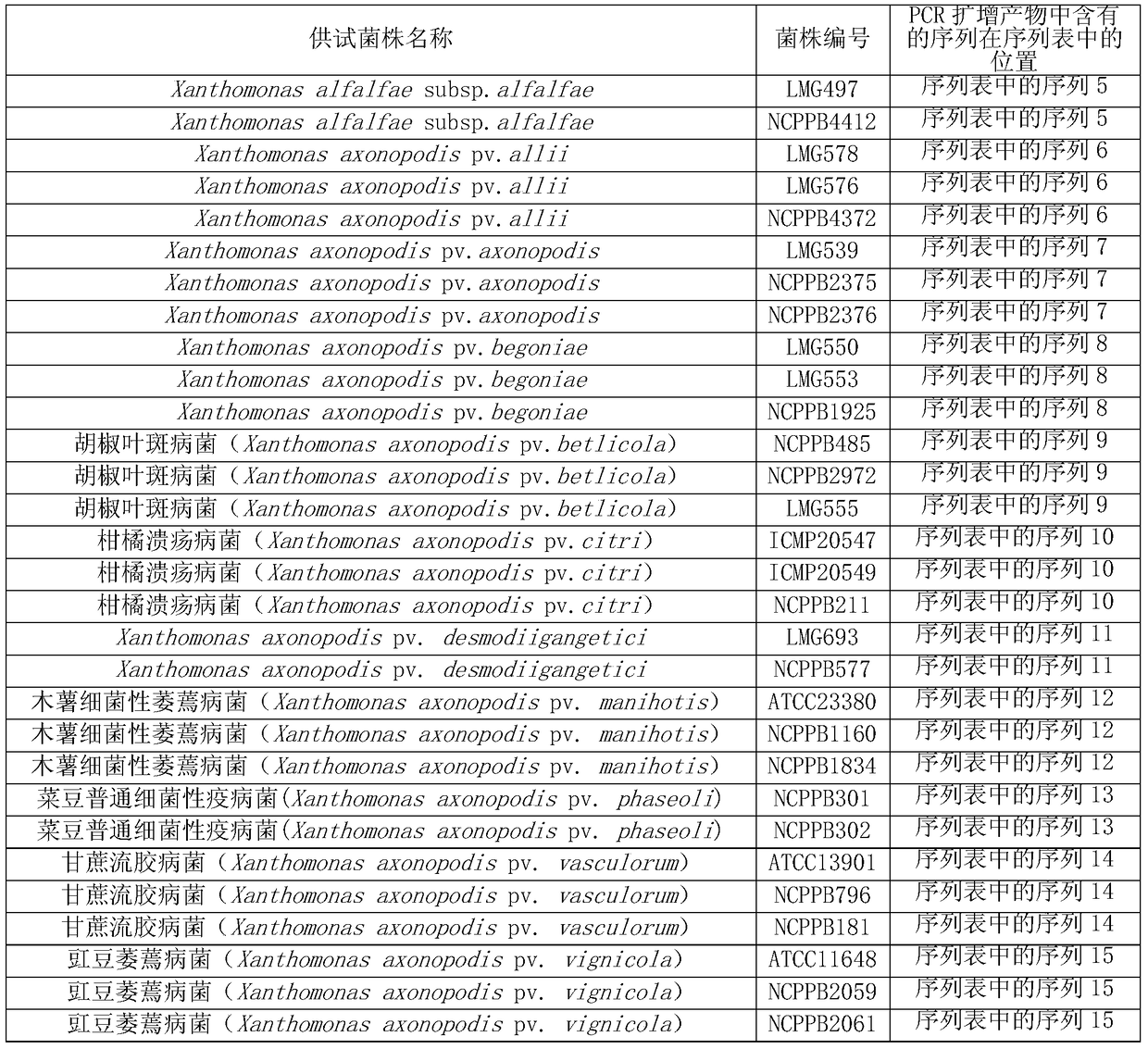
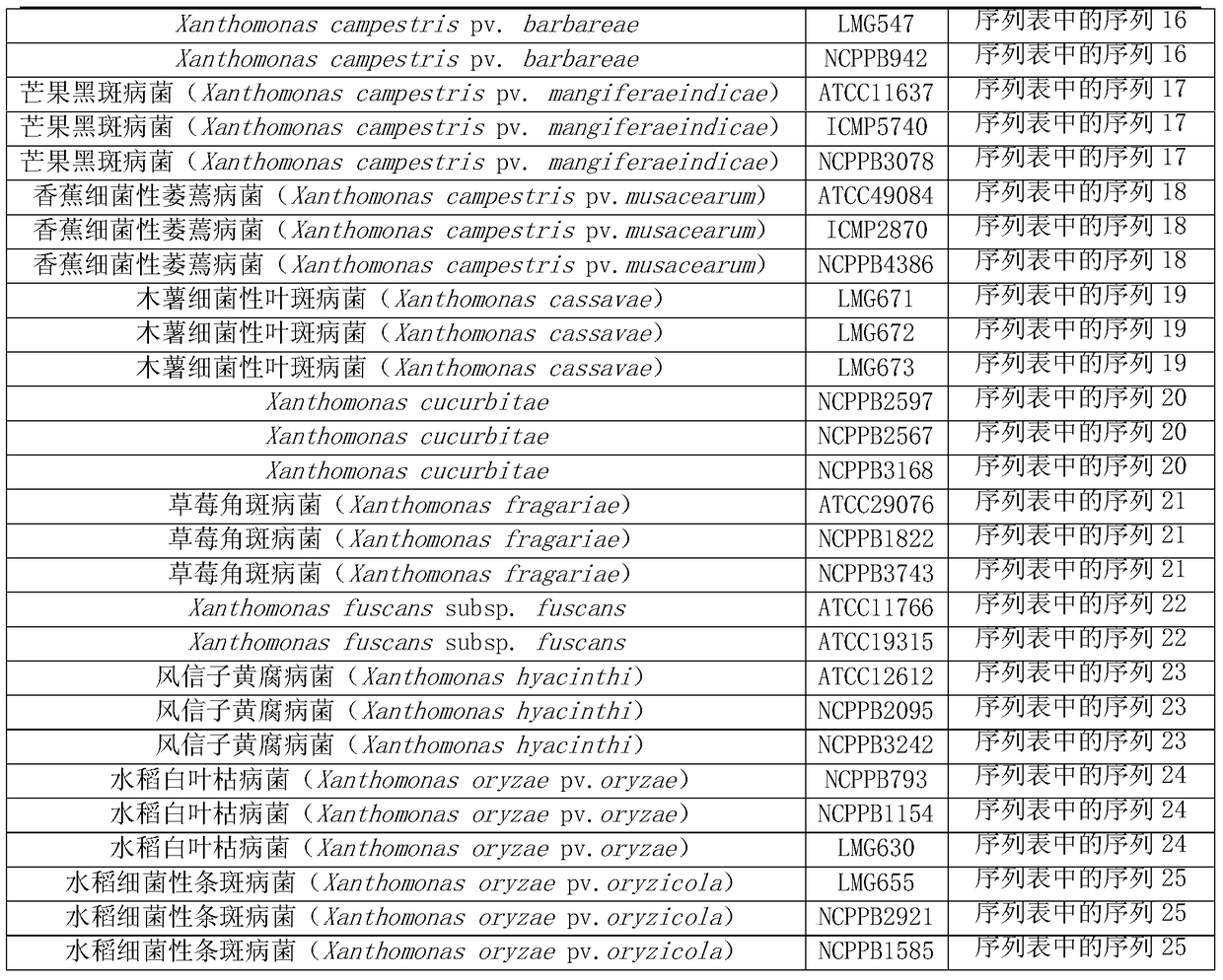
[0099] The basic information of the 58 tested strains is shown in Table 1. Among them, there were 11 kinds of quarantine Xanthomonas, 3 strains of each; 10 kinds of non-quarantine Xanthomonas, 25 strains.
[0100] The bacterial strain numbers LMG497, LMG578, LMG576, LMG539, LMG550, LMG553, LMG555, LMG693, LMG547, LMG671, LMG672, LMG673, LMG630 and LMG655 are deposited in BCCM / LMG BacteriaCollection (website: http: / / bccm.belspo.be / index.php ), the public can obtain from this repository.
[0101] 菌株编号为NCPPB4412、NCPPB4372、NCPPB2375、NCPPB2376、NCPPB1925、NCPPB485、NCPPB2972、NCPPB211、NCPPB577、NCPPB1160、NCPPB1834、NCPPB301、NCPPB302、NCPPB796、NCPPB181、NCPPB2059、NCPPB2061、NCPPB942、NCPPB3078、NCPPB4386、NCPPB2597、NCPPB2567、NCPPB3168、NCPPB1822 , NCPPB3743, NCPPB2095, NCPPB3242, NCPPB793, NCPPB1154, NCPPB2921 and NCPPB1585 are preserved in the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com