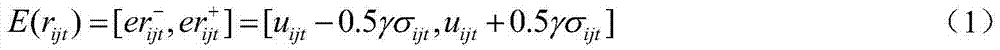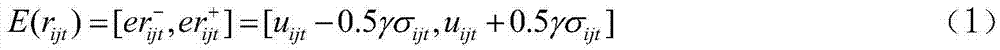Time sequence data processing method based on dynamic network diagram analysis
A time series, dynamic network technology, applied in electrical digital data processing, special data processing applications, instruments, etc., can solve problems such as small number of samples, large number of variables, and sparse time points.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
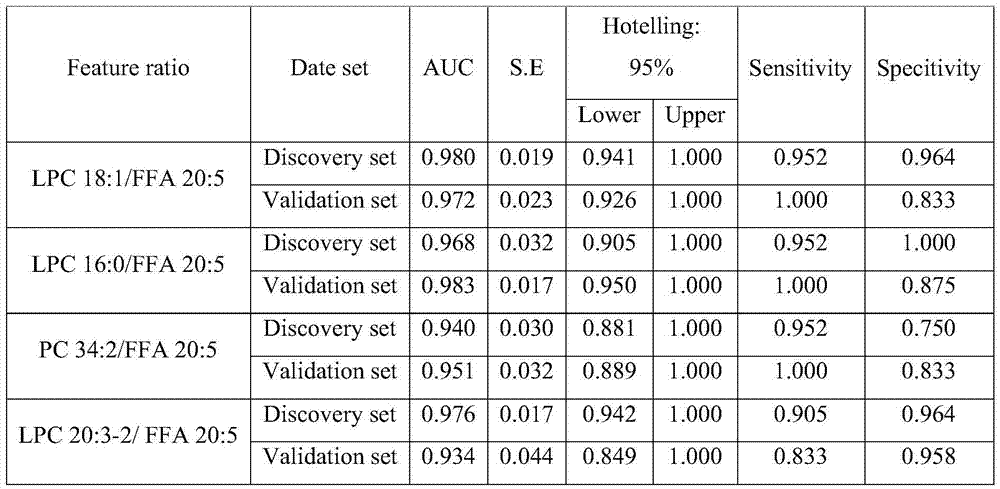
[0034] Example: Screening of liver disease early warning markers based on serum metabolic profile.
[0035] (1) Collection and pretreatment of rat serum samples.
[0036] Diethylnitrosamine was used to induce progressive carcinogenesis in rats. The discovery set contained 10 rats in the control group and 7 rats in the model group. From week 8 (T 1 ) to week 20 (T 7 ), serum samples were collected every 2 weeks, and a total of 119 serum samples were collected at 7 time points. In addition, this biological experiment also included an independent test set consisting of another 6 model group rats. At the 18th week, the liver tissues of these 6 rats were taken for histological examination to determine whether cancer occurred. Therefore, the test set contains 6 time points and a total of 36 serum samples.
[0037] (2) There are 3 liver diseases in the discovery set (N s = 3), T 1 It is a typical hepatitis stage (H), T 2 to T 4 For the stage of liver cirrhosis, T 5 to T 7...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com