Specific gene of brown atrichopogon and molecular identification method of brown atrichopogon
A specific gene and molecular identification technology, applied in the field of species identification, can solve problems such as difficulty, long cycle, and cumbersome operation steps, and achieve the effect of shortening identification time and ensuring accuracy and reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
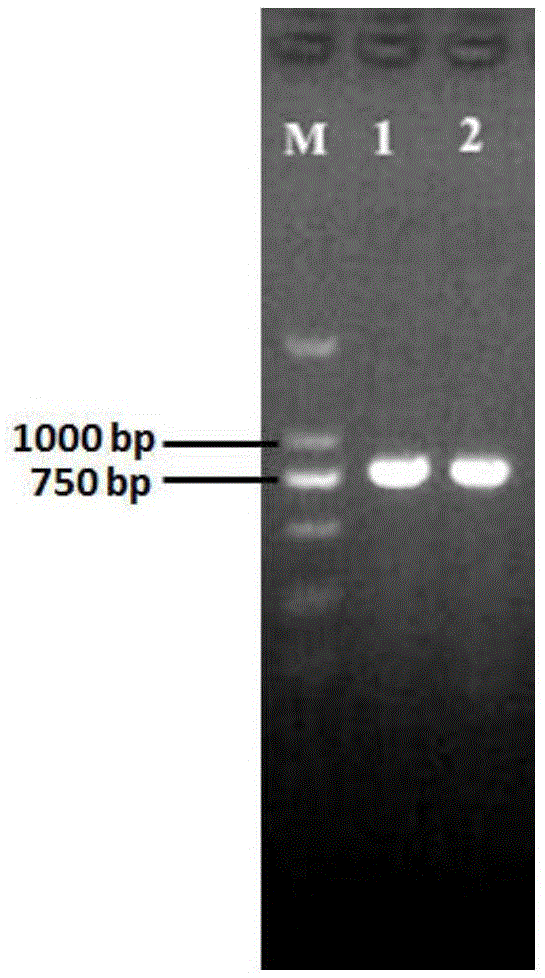
Image
Examples
Embodiment 1
[0024] Example 1 Acquisition of 18SrDNA gene sequence of Atrichopogon subfusculus
[0025] 1. Acquisition of the specimens of the subpalatin midge
[0026] The subpalatinous midges were collected from the field, and they were killed by freezing and stored in 95% ethanol directly. After the specimens are made into slides, they are identified by authoritative experts to ensure the accuracy of the identification results. Before making specimens, pick the end of the thorax and the first few segments of the abdomen of the sub-brown midge and place them in 95% alcohol for DNA extraction.
[0027] 2. DNA template preparation
[0028] Using the small insect DNA template preparation method to extract the DNA of the subpalatin midge (Edited by Wen Lizhang. Introduction to Entomological Research Methods and Techniques [M]. Beijing: Science Press, 2010.pp278-286), the extracted DNA samples are stored in Standby at -20°C.
[0029] 3. Primer synthesis
[0030] The primers used in this ...
Embodiment 2
[0045] The identification of embodiment 2 unknown naked midges
[0046] 1. Collection and preservation of specimens
[0047] The collected specimens were collected by waving nets or lamp lures, and the collected specimens were killed by freezing and stored in 95% ethanol directly.
[0048] 2. DNA template preparation
[0049] Under a dissecting mirror or a stereo microscope, a female nude midge was randomly selected from the collected midge specimens, and the DNA of the nude midge was extracted by using the small insect DNA template preparation method (Wen Lizhang, editor-in-chief. Entomological research methods and techniques Introduction [M]. Beijing: Science Press, 2010.pp278-286), the extracted DNA samples were stored at -20°C for future use, and each sample had a number.
[0050] 3. Acquisition of target gene sequence
[0051] 3.1 Primer synthesis
[0052] The obtained unknown naked midge DNA was used as a template to synthesize primers, and the sequences of the prime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com