Double droplet type digital PCR absolute quantification detection kit for AIV H7N9 subtypes
A detection kit, H7N9 technology, applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of easy mutation of bird flu virus, lagging of AIVH7N9 detection work, etc., to achieve high accuracy, Easy to operate and highly specific effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
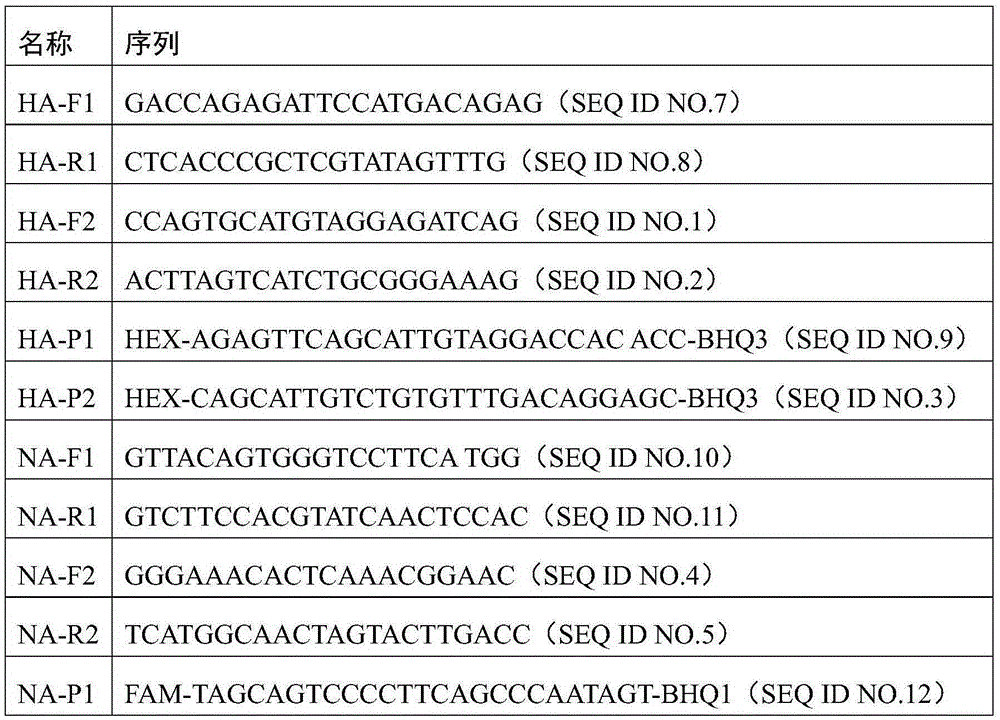
[0040] Embodiment 1 detects the design of the specific primer and probe of avian influenza H7N9 subtype
[0041] 1. Design of primers and probes of the present invention: the hemagglutinin gene (HA gene, GenBank accession number is KP417119) of avian influenza H7N9 (AIV-H7N9) as the target gene, and the nerve gene of avian influenza H7N9 (AIV-H7N9) The amino acid enzyme gene (NA gene, GenBank accession number KR351266) was used as the target gene, and specific primers suitable for ddPCR were designed.
[0042] This example gives the process of screening the best primers, select several pairs of alternative primers designed by software for screening, the alternative primers are as follows, see Table 1.
[0043] Table 1
[0044]
[0045]
[0046] 2. Screening of primers
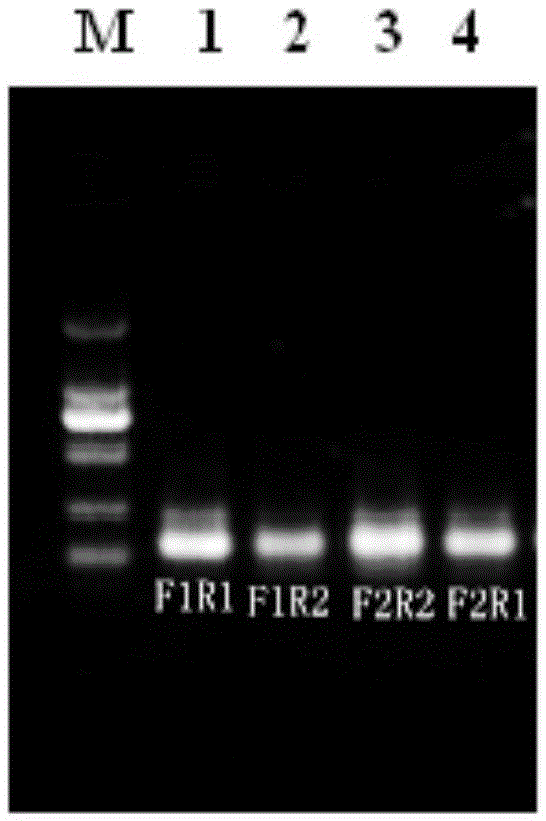
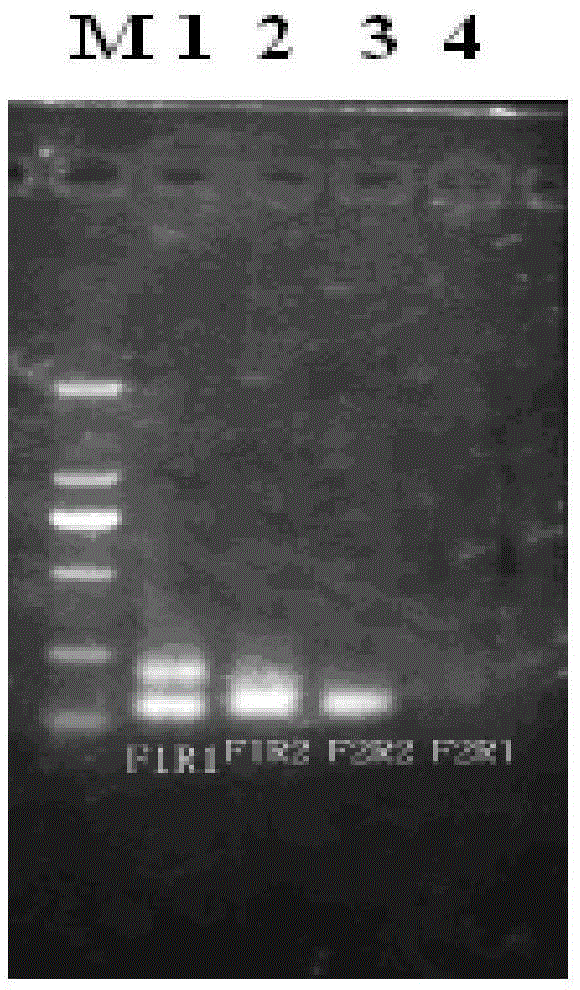
[0047] (1) The HA primers were randomly matched into four pairs: F1R1, F1R2, F2R1, F2R2; the NA primers were randomly matched into four pairs: F1R1, F1R2, F2R1, F2R2.
[0048] (2) Then reverse transcri...
Embodiment 2
[0055] Example 2 Optimization of the annealing temperature of the PCR method for detecting H7N9 on the ddPCR platform.
[0056] 1. Select the combination of primers and probes as: HA-F2R2P2+NA-F2R2P2.
[0057] 2. Then on the ddPCR platform, ddPCR system formula: 2XOne-stepRT-ddPCRSupermix 10μl, upstream primer, downstream primer 1μl, probe 0.5μl, RNaseFreedH 2 O3μl, positive template 2μl, total volume 20μl (concentration of both primers and probes is 10μM). Do 8 replicate wells and generate microdroplets.
[0058] 3. Amplify on a PCR instrument. The PCR amplification program is reverse transcription at 50°C for 10 minutes; pre-denaturation at 95°C for 10 minutes; denaturation at 94°C for 30 sec, and annealing at 50-60°C (8 temperature gradients are automatically distributed by the instrument) for 60 sec, a total of 40 sec. cycle; 98 ° C for 10 min to end the reaction. Detection on the droplet digital PCR detector.
[0059] 4. Analyzing results: Select an annealing temperat...
Embodiment 3
[0060] The optimization experiment of embodiment 3 primers, probe concentration
[0061] The concentrations of the designed primers and probes were all 10 μM, and the combination was HA-F2R2P2+NA-F2R2P2. The system configuration method is shown in Table 2.
[0062] Table 2
[0063]
[0064]
[0065] Then micro-droplets were generated on the ddPCR platform, transferred to a 96-well plate, and sealed with an aluminum film.
[0066] Amplify on a PCR instrument. The amplification program of PCR is reverse transcription at 50°C for 10 minutes; pre-denaturation at 95°C for 10 minutes; denaturation at 94°C for 30 seconds, annealing at 55°C for 60 seconds, a total of 40 cycles; and 98°C for 10 minutes to end the reaction. Detection on the droplet digital PCR detector.
[0067] Analysis results: According to the experimental results, the formula of HA-F2R2P2+NA-F2R2P2 primers and probes was selected, and the selected primers were 1.8 μl and the probes were 0.4 μl respectively. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com