PCR (polymerase chain reaction) primer group for species identification of cannabins satival, boehmeria nivea and linum usitatissimum fibers and PCR identification method
A primer set and ramie technology are applied in the PCR primer set for identifying hemp, ramie, and flax raw hemp fiber types, and in the field of PCR identification, which can solve problems such as difficulties and achieve the effects of reliable results, small sample size, and convenient detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029]Embodiment 1: primer set described in the present invention
[0030] Hemp detection upstream and downstream primers: see SEQ ID NO:1 for the upstream primer, specifically 5′-ATCGAAGAGCAGCAAACAAGC-3′, see SEQ ID NO:2 for the downstream primer, specifically 5′-TGGAGATCTTGTCTTCCAGCTG-3′;
[0031] Ramie detection upstream and downstream primers: see SEQIDNO:3 for the upstream primer, specifically 5'-TCCAAGCATTTCCCGAACGA-3', see SEQIDNO:4 for the downstream primer, specifically 5'-GCCCGATCTTCCAGCTCTAC-3';
[0032] Flax detection upstream and downstream primers: see SEQ ID NO:5 for the upstream primer, specifically 5′-TCATCAACAGCGCATCTGGT-3′, and refer to SEQ ID NO:6 for the downstream primer, specifically 5′-AGCCTTACAGCCACACACTC-3′.
Embodiment 2
[0033] Embodiment 2: PCR identification method of the present invention
[0034] Adopt commercially available commercial DNA extraction kit to extract the DNA of the hemp plant fiber sample to be tested;
[0035] Adopt the nucleotide sequence shown in SEQIDNO:1-2 or the nucleotide sequence shown in SEQIDNO:3-4 or the nucleotide sequence shown in SEQIDNO:5-6 to carry out PCR amplification to sample DNA;
[0036] The PCR amplification system is:
[0037] The total reaction volume is 25μl, including 20ng sample DNA, 12.5μl 2×PremixTaq buffer, 1μl each of 5μM upstream and downstream primers, and the balance is ddH 2 O.
[0038] The procedure for PCR amplification is:
[0039] Pre-denaturation at 95°C for 5 minutes;
[0040] Denaturation at 95°C for 30s, annealing at 55°C for 30s, extension at 72°C for 45s, cycled 27 times;
[0041] Extend at 72°C for 10min, and store at 10°C.
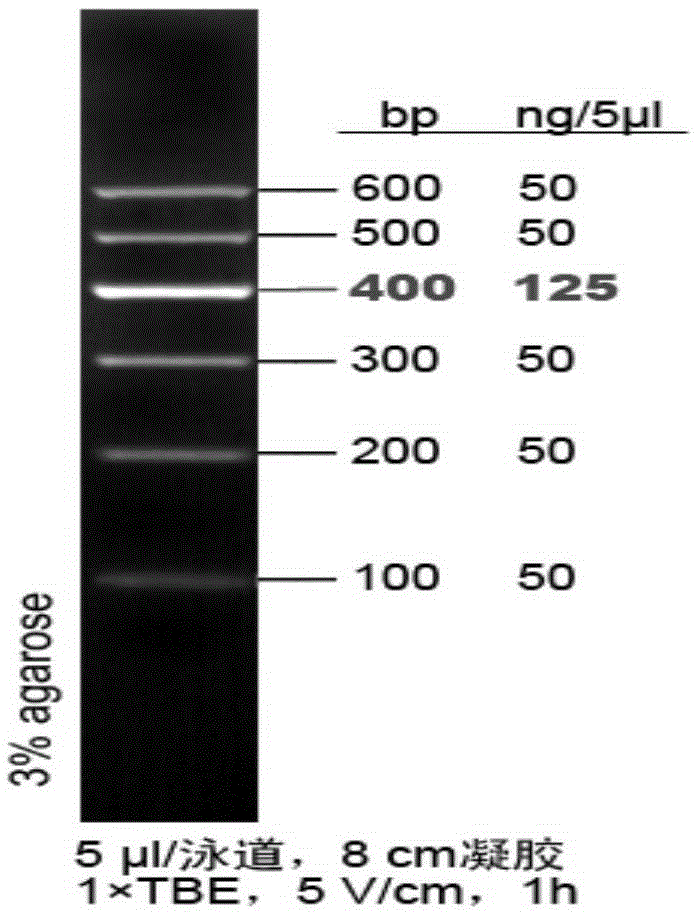
[0042] PCR amplified product carries out agarose gel electrophoresis (gel concentration 2.0%), if ...
Embodiment 3
[0043] Embodiment 3: identification test
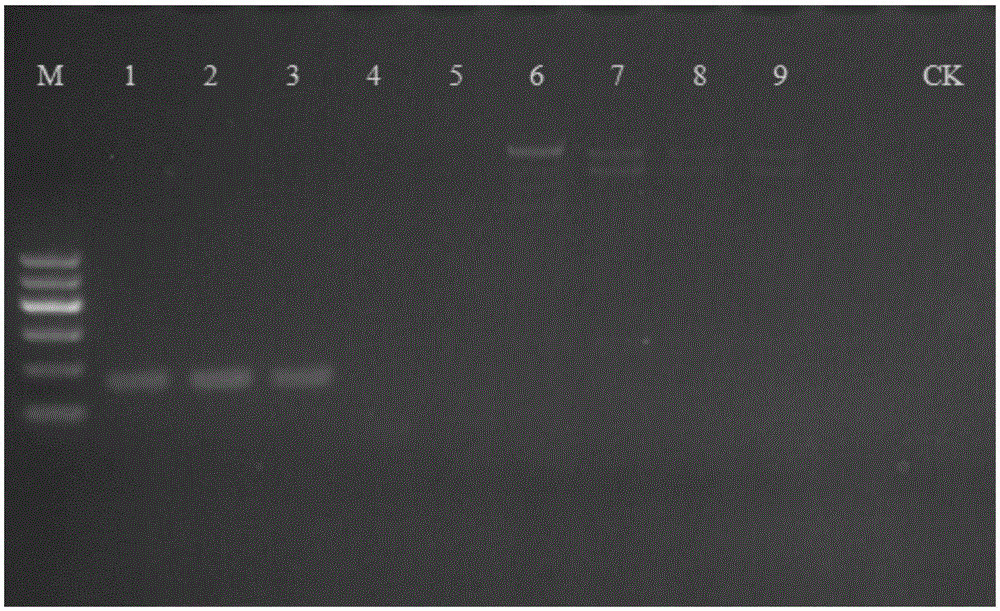
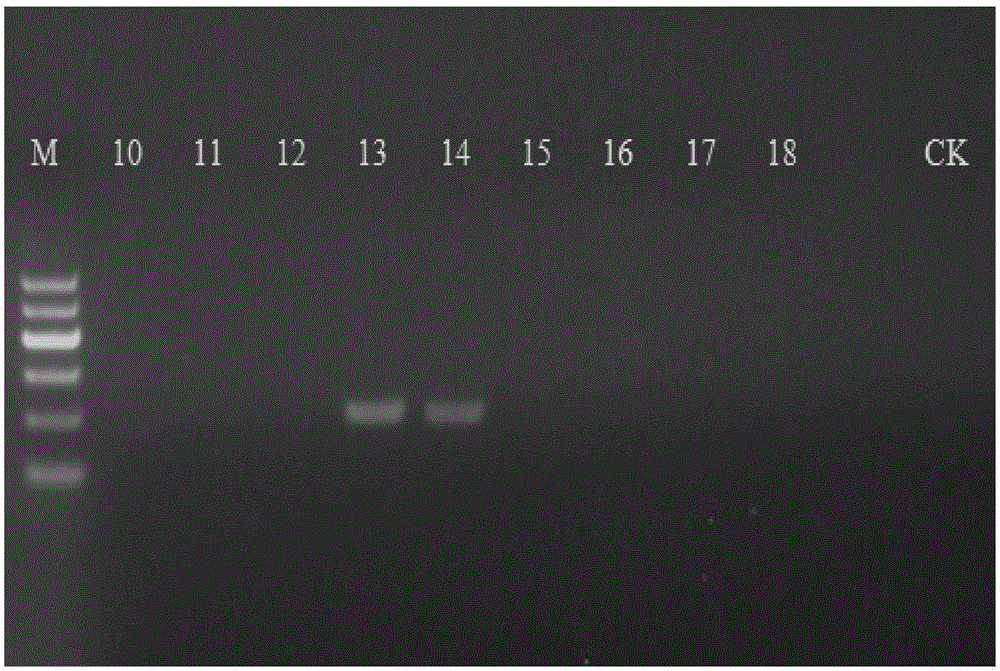
[0044] The test samples were hemp, ramie and flax, and the specific varieties and collection places are shown in Table 1.
[0045] Table 1 Test sample
[0046] sample number
breed name
collection place
A (hemp)
Yunma No.1
Xishuangbanna, Yunnan Province
B (hemp)
Heilongjiang industrial hemp
Mohe, Heilongjiang Province
C (hemp)
Anhui Cannabis
Lu'an, Anhui Province
D (ramie)
Xiangzhu No.2
Changsha, Hunan
E (ramie)
Xiangzhu No.3
Changsha, Hunan
F (flax)
West Asia One
Guyuan, Ningxia Province
G (linen)
Gia II
Changchun City, Jilin Province
H (linen)
Gia IV
Changchun City, Jilin Province
I (linen)
Gia Six
Changchun City, Jilin Province
[0047] Adopt the nucleotide sequence shown in SEQIDNO:1-2 to carry out PCR amplification to above-mentioned sample DNA respectively and carry ou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com