Method for amplification and assay of RNA fusion gene variants, method of distinguishing same and related primers, probes, and kits
A fusion and gene technology, applied in the field of oligonucleotide primers and probes, kits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0193] This example describes the design of primers for BCR-ABL amplification.
[0194] Amplification involved the use of forward and reverse primers positioned on opposite sides of the translocation breakpoint between the fused BCR and ABL gene segments. Thus, each resulting BCR-ABL amplicon contained a variant-specific linker sequence. One reverse primer (RP) and three forward primers (FP) were prepared. The reverse primer a2RP anneals to a specific sequence within exon a2 of ABL that is shared by variants e1a2, b2a2, b3a2 and e19a2. The reverse primer a2RP directs reverse transcription of RNA from multiple variants. Forward primers e1FP, b2FP and e19FP anneal to specific sequences within exons e1, b2 and e19 of the BCR, respectively. The primer pair e1FP and a2RP will amplify the cDNA target sequence reverse transcribed from the variant e1a2 RNA. The primer pair b2FP and a2RP will amplify the cDNA target sequence reverse transcribed from the variant b2a2 and b3a2 RNA. ...
Embodiment 2
[0204] This example describes the design of the BCR-ABL probe.
[0205]Four short oligonucleotide probes were designed with high affinity to junction-specific sequence specificity from each of the targeted BCR-ABL variants (i.e., e1a2, b2a2, b3a2, and e19a2) to hybridize. The BCR-ABL exon junctions spanned by each probe are separated by intron sequences in the genomic DNA. Thus, the probe hybridizes only to amplicons derived from BCR-ABL RNA / cDNA. The specificity of the probe will enable discrimination of the variants. The probes can be used individually to detect a single BCR-ABL variant. Alternatively, two or more probes can be used together to detect multiple BCR-ABL variants in a single reaction. Probes can be labeled identically for combined detection of targeted BCR-ABL variants. Alternatively, for differential detection of two or more BCR-ABL variants in a single reaction, the probes can be labeled differently. The sequences of the probes are shown in Table IV.
...
Embodiment 3
[0224] This example describes the design of internal control (IC) amplification primers and probes.
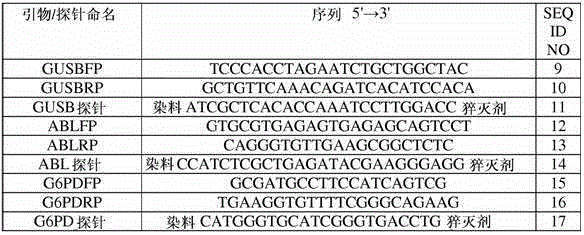
[0225] Two oligonucleotide primers (ie, forward primer and reverse primer) were designed for amplification of each of the three endogenous genes used as internal controls. The primers were designed to amplify the well-characterized housekeeping genes β-glucuronidase (GUSB), ABL and glucose-6-phosphate dehydrogenase (G6PD).
[0226] The GUSB forward primer (GUSBFP) anneals to a sequence located in exon 8 of GUSB. The GUSB reverse primer (GUSBRP) anneals to a sequence located in exon 10 of GUSB, and in combination with GUSBFP directs the amplification of the reverse transcribed GUSB cDNA. The resulting amplicon spans the sequence from exons 8, 9 and 10 of GUSB.
[0227] Oligonucleotide probes were designed for hybridization and detection of GUSB amplicons. The probe was designed to hybridize to sequences in exon 9 of GUSB.
[0228] The ABL forward primer (ABLFP) anneals to a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com