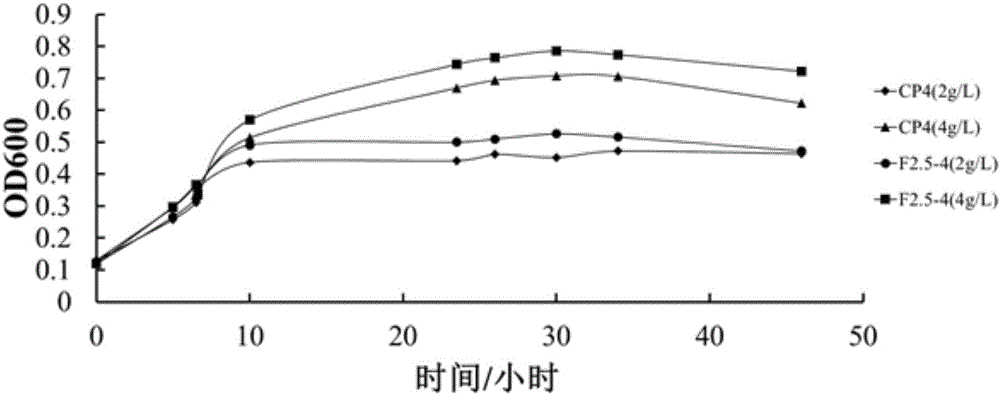
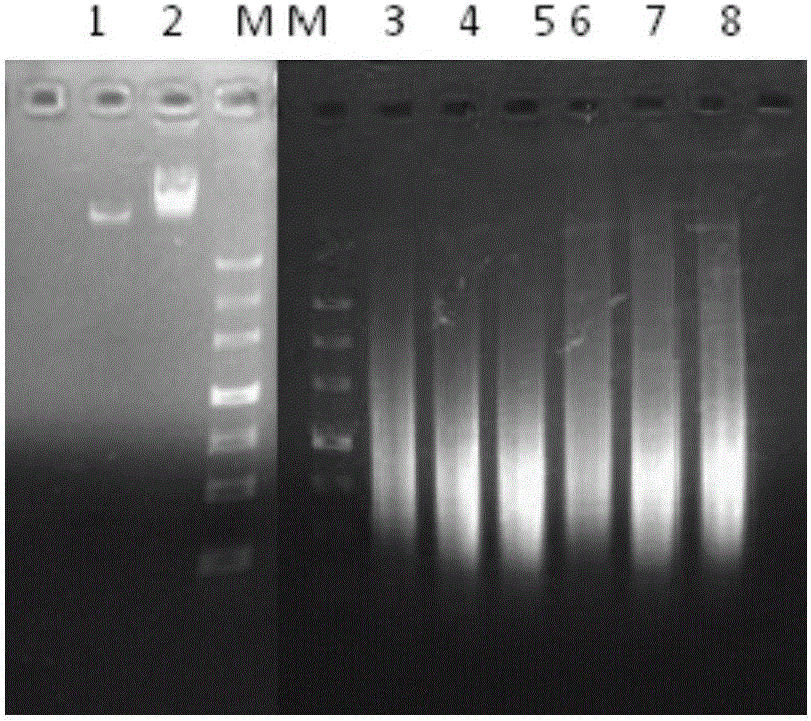
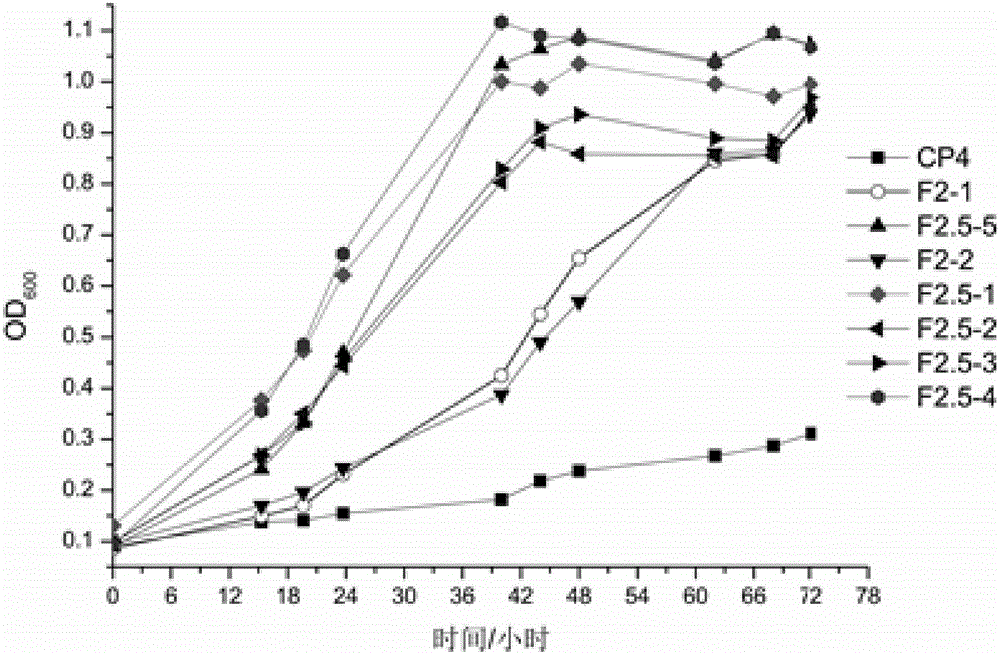
Error-prone whole-genome shuffling method of zymomonas mobilis and furfural-tolerant zymomonas mobilis
An error-prone whole-genome, Zymomonas technology, applied in the field of bioengineering, to achieve the effect of solving the energy crisis, strong tolerance to furfural, fast and efficient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Genomic DNA extraction:
[0035] Extract the genomic DNA of the starting strain Zymomonas mobilis CP4, ( figure 1 ), the specific operation is as follows: the above-mentioned bacterial liquid in the glycerol tube is transferred to a test tube containing 5 mL of RM liquid medium, cultured at 30°C overnight, and OD600=2~4. The culture was transferred to a centrifuge tube, centrifuged at 13000 rpm at 4°C for 1 minute, and then the supernatant was discarded. After the pellet was resuspended in 30 μL of sterile water, the supernatant was discarded by centrifugation. Add 120μL of bactericidal buffer, 60μLTris saturated phenol, 60μL of chloroform, 120μLTE buffer, 120μL volume of quartz sand into the centrifuge tube, shake well for 4min, centrifuge at 4℃13000rpm / 5min to get the supernatant. Add 1 mL of absolute ethanol to the supernatant, mix upside down, place at -20°C for 30 minutes, centrifuge at 4°C, 13000rpm / 25min, discard the supernatant, dry the pellet, and take 5...
Embodiment 2
[0041] Example 2 Error-prone PCR amplification of the whole genome:
[0042] Take 5μL of dNTPS stock solution, 16.6μL of 100uM10-17 base random primer, 3μL of 15mM MnCl 2 , 5μL of 10× mutation buffer, 20ng of genomic DNA obtained in step (1), 2.5μL of 2U / μLTaqDNA polymerase, supplemented with sterile water to 50μL; the dNTPS mother liquor contains 10mM dCTP and dTTP, 2mMdATP and dGTP; the 10 ×Mutant buffer contains 1mM Tris-HCl pH8.3, 0.7mM MgCl 2 , 5mMKCl, 1g / 100mL glycerol;
[0043] Error-prone PCR reaction conditions: 94°C pre-denaturation for 5 minutes; 92°C for 1 minute, then annealing at 37°C for 1 minute, then from 37°C to 55°C at a rate of 0.1°C per second, and 55°C extension for 4 minutes. 50 cycles, supplementary extension at 55℃ for 10min; amplification products such as figure 1 Shown.
[0044] 16.6μL of 100uM10-17 bases of any primer can amplify error-prone PCR products.
[0045] 16.6μL 100uM10-17 bases are:
[0046] 8.3μL100uM10 base random primer and 8.3μL100uM12base ran...
Embodiment 3
[0060] Example 3 Whole genome reorganization:
[0061] Mix the various PCR products obtained in Example 2, and concentrate them by ethanol precipitation 10 times for electroconversion;
[0062] After the starting strain Zymomonas mobilis CP4 was inoculated into 3ml RM liquid medium and grown to the late logarithmic stage, its culture was inoculated into 50ml RM liquid medium with 0.5% inoculum, and cultivated at 30℃ to OD. 600 = 0.4-0.5 (it can also be any value of 0.4-0.5); centrifuge at 6000 rpm for 5 min, collect the bacteria, wash twice with an ice bath of 10% glycerol aqueous solution with a volume percentage of 2ml Suspend and mix in 10% glycerol aqueous solution to obtain competent cells;
[0063] Take 4 aliquots of competent cells, each 50μl, add 10 times concentrated PCR product 3μL, 4μL, 5μL, and 5μL sterile ultrapure water to 50μl competent cells, respectively transfer to sterile pre-cooled 1mm electric shock In the cup, 1800V electric shock, add 450ulRM liquid medium pre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com