Establishment and application of plant multi-gene knockout vector
A technology for knocking out vectors and multiple genes, which can be used in the introduction of foreign genetic material using vectors, recombinant DNA technology, etc., which can solve the problems of time-consuming, labor-intensive, and inability to obtain multiple mutants.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
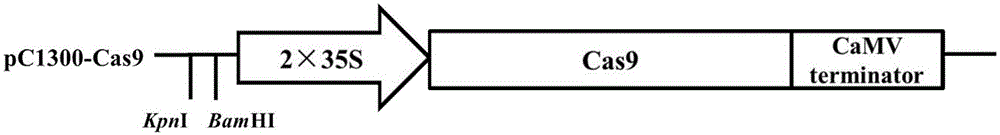
[0042] Example 1, construction of binary expression vector pC1300-Cas9
[0043] With primer pair 35S-F:5'-AAA AAA GTACCCCTACTCCAAAAATG-3'( labeled as KpnI, Marked as BamHI restriction site) and 35S-R:5'-ACA AGCTTGGGCTGTCCTCTCC-3'( Marked as the BglII restriction site) PCR amplification of plasmid pJIT163-2NLSCas9 (provided by the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), KODFX DNA polymerase (purchased from Toyobo Biotechnology Co., Ltd.) PCR amplification of the target band, reaction conditions as follows:
[0044]
[0045] Reaction program: denaturation at 94°C for 2 minutes; then denaturation at 98°C for 10 seconds, annealing at 58°C for 30 seconds, extension at 68°C for 40 seconds, and 31 cycles of amplification; finally extension at 68°C for 5 minutes.
[0046] A constitutive 2×CaMV35S promoter fragment of about 740bp was obtained, which was double-digested with KpnI and BglII and then ligated into the KpnI and BamHI rec...
Embodiment 2
[0047] Embodiment 2, the construction of intermediate carrier SK-gRNA
[0048] Use primer pair gRNA-F:5'-TTG GA CTCGAG GATTATGTGGAAAAAAAAGCACC-3'( Marked as KpnI restriction site, Marked as BamHI restriction site, ____ is marked as XbaI restriction site, _ is marked as XhoI restriction site) and gRNA-R:5'-CCGCGGTG GCTAGC CGATTAAGGAATCT-3'( Marked as the NotI restriction site, Marked as BglII restriction site, _ marked as NheI restriction site, ____ marked as SalI restriction site) PCR amplification plasmid pU3-gRNA (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences); KODFX DNA polymerase ( Purchased from Toyobo Biotechnology Co., Ltd.) PCR amplification target band, the reaction conditions are as follows:
[0049]
[0050] Reaction program: denaturation at 94°C for 2 minutes; then denaturation at 98°C for 10 seconds, annealing at 58°C for 30 seconds, extension at 68°C for 30 seconds, and 31 cycles of amplification; finally exten...
Embodiment 3
[0055] Example 3, Selection of target sequence in monocot rice and construction of a single gRNA intermediate vector
[0056] In the present invention, four genes of OsPDS, Os02g23823, OsMPK2 and Os05g02070 are simultaneously knocked out as an example, but not limited thereto.
[0057] 1. Selection of target sequence and primer design
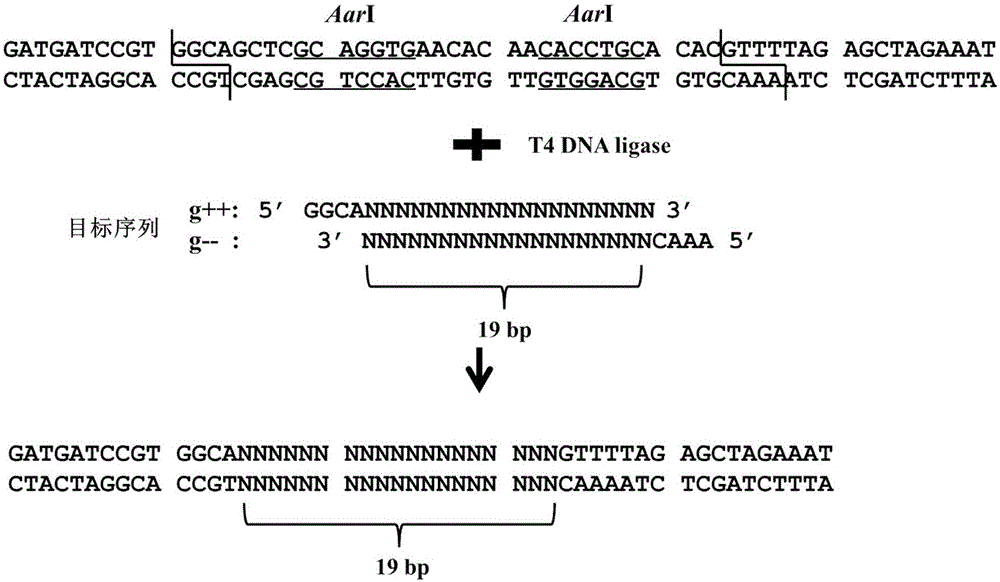
[0058] Find on the genome sequence of the target gene NNNNNNNNNNNNNNNN↓NNN The sequence is the target sequence (single underscore indicates the target sequence, double underscore indicates the PAM sequence, ↓ indicates the proposed cutting mutation site of CAS9 protein, at the first 3 bases of PAM.) Design two complementary DNA sequences: in the forward direction Add GGCA before the sequence and AAAC before the reverse complementary sequence. The specific structure is as image 3 shown.
[0059] details as follows:
[0060] Target gene: OsPDS, gene accession number: LOC_Os03g08570.1, target sequence: TTGGTCTTTGCTCCTG↓CAG (The double u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com