Preparation method of sea-buckthorn seed polypeptide used for sobering up from drunkenness
A technique for seabuckthorn seeds and sobering up, which is applied in the field of preparation of seabuckthorn seed polypeptides, can solve problems such as no research report on seabuckthorn seed dregs sobering polypeptides, and achieve the effect of high polypeptide yield and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
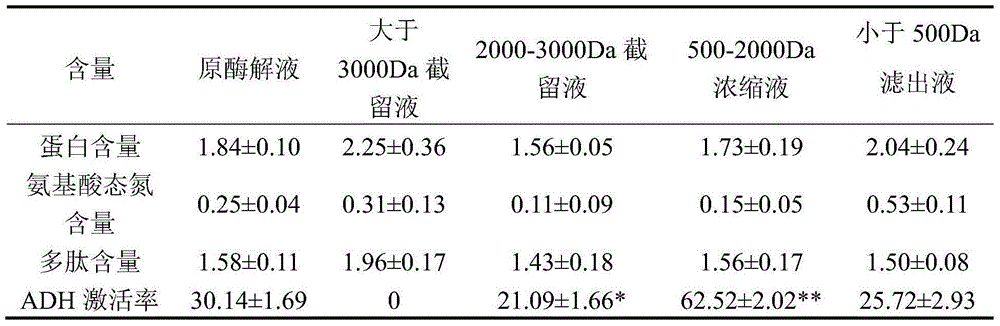
[0014] Take 50g of pulverized defatted seabuckthorn seed dregs (26.25% protein content), add 800ml of distilled water, place it at room temperature for 10 hours, adjust the pH to 6.5, add 0.06g of papain and hydrolyze it at 60°C for 2.5h, after the enzyme is inactivated The enzymolysis solution was centrifuged at 5000r / min to obtain the supernatant, which was concentrated until the polypeptide content reached 1.5% for later use. After microfiltration, the enzymatic hydrolyzate of seabuckthorn seeds was used to determine the activation rate of alcohol dehydrogenase by Waller-Hoch method. After determination, when the polypeptide content is 15.8mg / mL, the activation rate of the polypeptide solution to alcohol dehydrogenase is 30.14%. Two components with molecular mass Mw>3000Da and Mw3000Da had no activation rate of alcohol dehydrogenase, and the component with Mw<3000Da had no effect on ethanol dehydrogenase The activation rate of hydrogenase was 57.41%, significantly higher t...
Embodiment 2
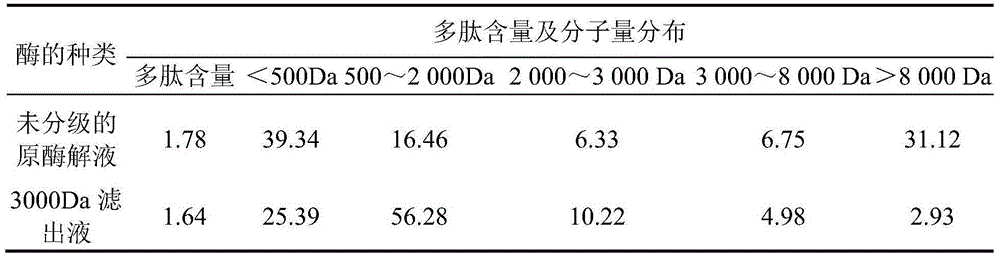
[0016] Take 50g of crushed defatted seabuckthorn seed dregs, add 600ml of distilled water, let it stand at room temperature for 8 hours, adjust the pH to 7.0, add 0.07g of papain and hydrolyze it at 50°C for 4.5h, inactivate the enzymatic solution at 5000r / The supernatant was collected by centrifugation for 1 min, and concentrated until the peptide content reached 1.5%. After microfiltration of the enzymatic solution, select ultrafiltration membranes and nanofiltration membranes with molecular weight cut-offs of 3000Da, 2000Da and 500Da to separate and obtain 4 components, namely molecular weight Mw>3000Da, Mw between 2000 and 3000Da, and Mw between 500 and 500Da. 2000Da, Mw<500Da. After the 4 kinds of separation liquid and the original enzymatic hydrolysis liquid were evaporated and concentrated to the same polypeptide content, the activation rate of different samples to alcohol dehydrogenase was determined by Waller-Hoch method. Gel chromatography was used to analyze the m...
Embodiment 3
[0023] Take 50g of pulverized defatted seabuckthorn seed dregs, add 1000ml of distilled water, let it stand at room temperature for 10 hours, adjust the pH to 6.0, add 0.09g of papain and hydrolyze it at 55°C for 3.5h, and inactivate the enzymatic solution at 5000r / The supernatant was collected by centrifugation for 1 min, concentrated until the polypeptide content reached 1.5%, and then separated and purified by an ultrafiltration membrane with a molecular weight cut-off of 3000Da to obtain a seabuckthorn seed polypeptide solution with a molecular weight less than 3000Da, which was concentrated until the polypeptide content reached 4% for later use.
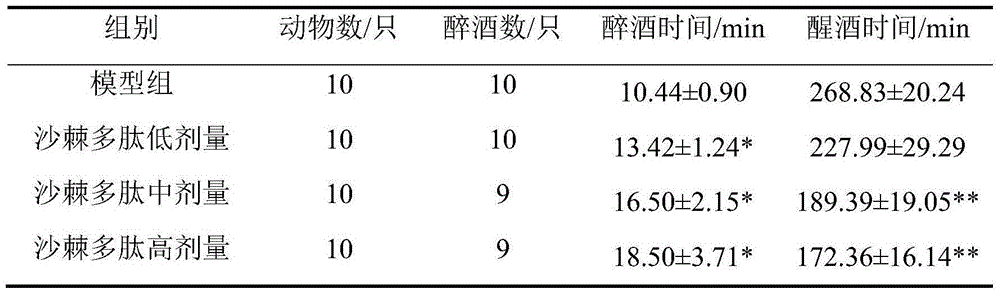
[0024] The anti-drunk test procedure is to pour the polypeptide sample first and then pour the drinking concentration at 56° of liquor. 40 male mice (half male and half male) were randomly divided into 4 groups: model group; low, medium and high dose (100, 200, 400 mg / kg·bw) groups of seabuckthorn polypeptide. The model group ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com